6IJJ
 
 | | Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X, Ma, J, Su, X, Liu, Z, Zhang, X, Li, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-03-20 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI supercomplex.
Nat Plants, 5, 2019
|
|
6IJO
 
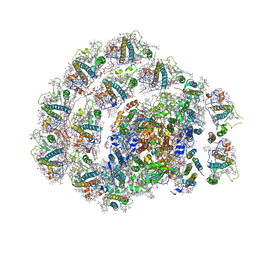 | | Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X, Ma, J, Su, X, Liu, Z, Zhang, X, Li, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-03-20 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI supercomplex.
Nat Plants, 5, 2019
|
|
5ZJI
 
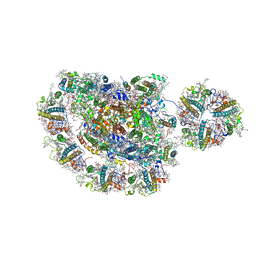 | | Structure of photosystem I supercomplex with light-harvesting complexes I and II | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Ma, J, Su, X.D, Cao, P, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-20 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the maize photosystem I supercomplex with light-harvesting complexes I and II.
Science, 360, 2018
|
|
6AGF
 
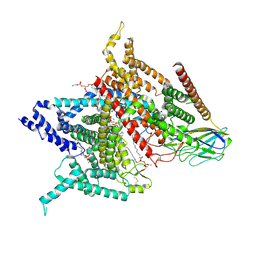 | | Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta1 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, X.J, li, Z.Q, Zhou, Q, Shen, H.Z, Wu, K, Huang, X.S, Chen, J.F, Zhang, J.R, Zhu, X.C, Lei, J.L, Xiong, W, Gong, H.P, Xiao, B.L, Yan, N. | | Deposit date: | 2018-08-11 | | Release date: | 2018-10-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta 1.
Science, 362, 2018
|
|
5YLX
 
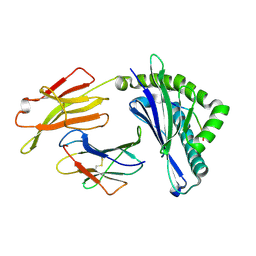 | | Integrated illustration of a valid epitope based on the SLA class I structure and tetramer technique could carry forward the development of molecular vaccine in swine species | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, PRRSV-NSP9-TMP9 peptide | | Authors: | Pan, X.C, Wei, X.H, Zhang, N, Xia, C. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2020-05-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Illumination of PRRSV Cytotoxic T Lymphocyte Epitopes by the Three-Dimensional Structure and Peptidome of Swine Lymphocyte Antigen Class I (SLA-I).
Front Immunol, 10, 2019
|
|
2G77
 
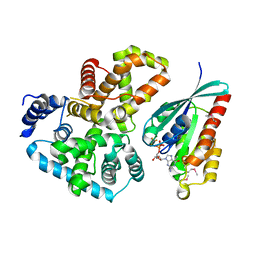 | | Crystal Structure of Gyp1 TBC domain in complex with Rab33 GTPase bound to GDP and AlF3 | | Descriptor: | ALUMINUM FLUORIDE, GTPase-activating protein GYP1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Pan, X, Eathiraj, S, Munson, M, Lambright, D.G. | | Deposit date: | 2006-02-27 | | Release date: | 2006-07-25 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism.
Nature, 442, 2006
|
|
3TOE
 
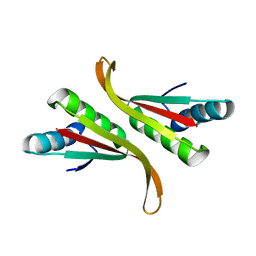 | | Structure of Mth10b | | Descriptor: | DNA/RNA-binding protein Alba | | Authors: | Pan, X.M, Zhang, N, Liu, Y.F, Liu, X. | | Deposit date: | 2011-09-05 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Molecular mechanism underlying the interaction of typical Sac10b family proteins with DNA.
Plos One, 7, 2012
|
|
5ZFL
 
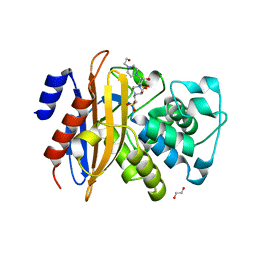 | | Crystal structure of beta-lactamase PenP mutant E166Y | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase | | Authors: | Pan, X, Zhao, Y. | | Deposit date: | 2018-03-06 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The hydrolytic water molecule of Class A beta-lactamase relies on the acyl-enzyme intermediate ES* for proper coordination and catalysis.
Sci Rep, 10, 2020
|
|
5ZG6
 
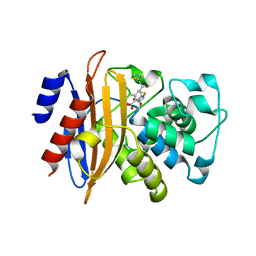 | |
5ZFT
 
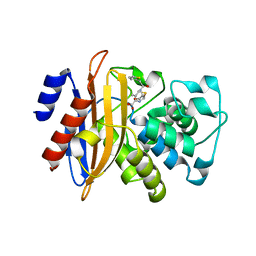 | |
6KHI
 
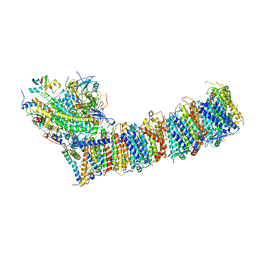 | | Supercomplex for cylic electron transport in cyanobacteria | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Pan, X, Cao, D, Xie, F, Zhang, X, Li, M. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase.
Nat Commun, 11, 2020
|
|
3PL9
 
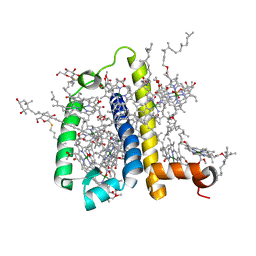 | | Crystal structure of spinach minor light-harvesting complex CP29 at 2.80 angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, M, Wan, T, Wang, L.F, Jia, C.J, Hou, Z.Q, Zhao, X.L, Zhang, J.P, Chang, W.R. | | Deposit date: | 2010-11-14 | | Release date: | 2011-02-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into energy regulation of light-harvesting complex CP29 from spinach.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1NDU
 
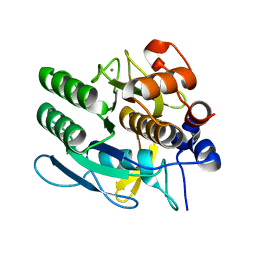 | |
1NDQ
 
 | | Bacillus lentus subtilisin | | Descriptor: | CALCIUM ION, Subtilisin Savinase | | Authors: | Pan, X, Bott, R, Glatz, C.E. | | Deposit date: | 2002-12-09 | | Release date: | 2004-04-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Subtilisin surface properties and crystal growth kinetics
J.CRYST.GROWTH, 254, 2003
|
|
6VGB
 
 | |
5XHR
 
 | | Crystal structure of P99 beta-lactamase in complex with a penicillin derivative MPC-1 | | Descriptor: | (2~{R},4~{S})-5,5-dimethyl-2-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-5-thiophen-2-yl-pentan-2-yl]-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase | | Authors: | Pan, X, Zhao, Y. | | Deposit date: | 2017-04-24 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modified Penicillin Molecule with Carbapenem-Like Stereochemistry Specifically Inhibits Class C beta-Lactamases
Antimicrob. Agents Chemother., 61, 2017
|
|
6VG7
 
 | |
6VGA
 
 | |
3JYN
 
 | | Crystal structures of Pseudomonas syringae pv. Tomato DC3000 quinone oxidoreductase complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Quinone oxidoreductase | | Authors: | Pan, X, Zhang, H, Gao, Y, Li, M, Chang, W. | | Deposit date: | 2009-09-22 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of Pseudomonas syringae pv. tomato DC3000 quinone oxidoreductase and its complex with NADPH
Biochem.Biophys.Res.Commun., 390, 2009
|
|
3JYL
 
 | | Crystal structures of Pseudomonas syringae pv. Tomato DC3000 quinone oxidoreductase | | Descriptor: | Quinone oxidoreductase | | Authors: | Pan, X, Zhang, H, Gao, Y, Li, M, Chang, W. | | Deposit date: | 2009-09-22 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Pseudomonas syringae pv. tomato DC3000 quinone oxidoreductase and its complex with NADPH
Biochem.Biophys.Res.Commun., 390, 2009
|
|
5GHY
 
 | |
8IJD
 
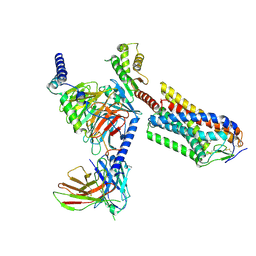 | | Cryo-EM structure of human HCAR2-Gi complex with MK-6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-27 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8IJA
 
 | | Cryo-EM structure of human HCAR2-Gi complex with niacin | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-26 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8IJ3
 
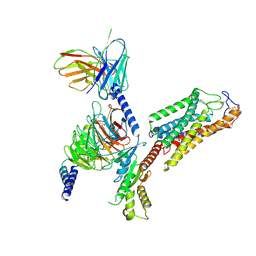 | | Cryo-EM structure of human HCAR2-Gi complex without ligand (apo state) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-24 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8IJB
 
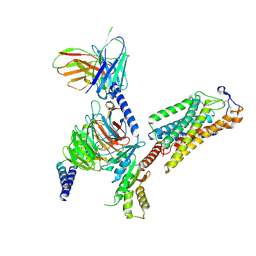 | | Cryo-EM structure of human HCAR2-Gi complex with acipimox | | Descriptor: | 5-methyl-4-oxidanyl-pyrazin-4-ium-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-27 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
