7DVO
 
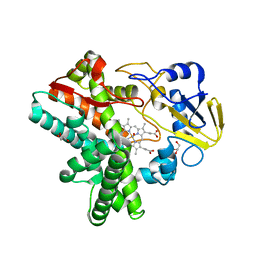 | | Structure of Reaction Intermediate of Cytochrome P450 NO Reductase (P450nor) Determined by XFEL | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Nomura, T, Kimura, T, Kanematsu, Y, Yamashita, K, Hirata, K, Ueno, G, Murakami, H, Hisano, T, Yamagiwa, R, Takeda, H, Gopalasingam, C, Yuki, K, Kousaka, R, Yanagasawa, S, Shoji, O, Kumasaka, T, Takano, Y, Ago, H, Yamamoto, M, Sugimoto, H, Tosha, T, Kubo, M, Shiro, Y. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Short-lived intermediate in N 2 O generation by P450 NO reductase captured by time-resolved IR spectroscopy and XFEL crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6JO2
 
 | | Ferredoxin I from Thermosynechococcus elongatus | | Descriptor: | BENZAMIDINE, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, ... | | Authors: | Motomura, T. | | Deposit date: | 2019-03-19 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An alternative plant-like cyanobacterial ferredoxin with unprecedented structural and functional properties.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
6IRI
 
 | |
5XCA
 
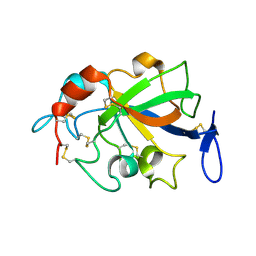 | | Crystal structure of GH45 endoglucanase EG27II D137A mutant in complex with cellobiose | | Descriptor: | Endo-beta-1,4-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nomura, T, Mizutani, K, Iwase, H, Takahashi, N, Mikami, B. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High-resolution crystal structures of the glycoside hydrolase family 45 endoglucanase EG27II from the snail Ampullaria crossean.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5XBX
 
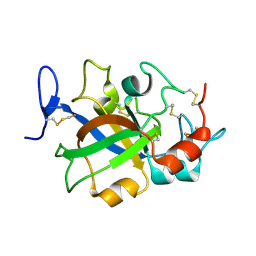 | | Crystal structure of GH45 endoglucanase EG27II in complex with cellobiose | | Descriptor: | Endo-beta-1,4-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nomura, T, Mizutani, K, Iwase, H, Takahashi, N, Mikami, B. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | High-resolution crystal structures of the glycoside hydrolase family 45 endoglucanase EG27II from the snail Ampullaria crossean.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5XC8
 
 | | Crystal structure of GH45 endoglucanase EG27II at pH5.5, in complex with cellobiose | | Descriptor: | Endo-beta-1,4-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nomura, T, Mizutani, K, Iwase, H, Takahashi, N, Mikami, B. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution crystal structures of the glycoside hydrolase family 45 endoglucanase EG27II from the snail Ampullaria crossean.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5XC4
 
 | | Crystal structure of GH45 endoglucanase EG27II at pH4.0, in complex with cellobiose | | Descriptor: | Endo-beta-1,4-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nomura, T, Mizutani, K, Iwase, H, Takahashi, N, Mikami, B. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High-resolution crystal structures of the glycoside hydrolase family 45 endoglucanase EG27II from the snail Ampullaria crossean.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5XC9
 
 | | Crystal structure of GH45 endoglucanase EG27II at pH8.0, in complex with cellobiose | | Descriptor: | Endo-beta-1,4-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nomura, T, Mizutani, K, Iwase, H, Takahashi, N, Mikami, B. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | High-resolution crystal structures of the glycoside hydrolase family 45 endoglucanase EG27II from the snail Ampullaria crossean.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5XBU
 
 | | Crystal structure of GH45 endoglucanase EG27II in apo-form | | Descriptor: | Endo-beta-1,4-glucanase | | Authors: | Nomura, T, Mizutani, K, Iwase, H, Takahashi, N, Mikami, B. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structures of the glycoside hydrolase family 45 endoglucanase EG27II from the snail Ampullaria crossean.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1UF2
 
 | | The Atomic Structure of Rice dwarf Virus (RDV) | | Descriptor: | Core protein P3, Outer capsid protein P8, Structural protein P7 | | Authors: | Nakagawa, A, Miyazaki, N, Taka, J, Naitow, H, Ogawa, A, Fujimoto, Z, Mizuno, H, Higashi, T, Watanabe, Y, Omura, T, Cheng, R.H, Tsukihara, T. | | Deposit date: | 2003-05-23 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins.
Structure, 11, 2003
|
|
2ZAH
 
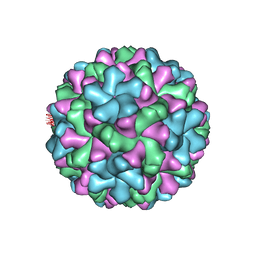 | | X-ray structure of Melon necrotic spot virus | | Descriptor: | CALCIUM ION, Coat protein, UNKNOWN ATOM OR ION | | Authors: | Wada, Y, Tsukihara, T, Omura, T. | | Deposit date: | 2007-10-05 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | X-ray structure of Melon necrotic spot virus
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3VJJ
 
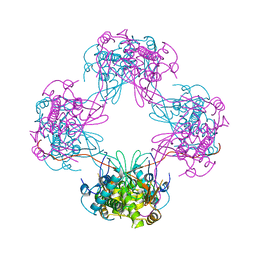 | | Crystal Structure Analysis of the P9-1 | | Descriptor: | P9-1 | | Authors: | Akita, F, Higashiura, A, Suzuki, M, Tsukihara, T, Nakagawa, A, Omura, T. | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analysis reveals octamerization of viroplasm matrix protein P9-1 of Rice black streaked dwarf virus
J.Virol., 86, 2012
|
|
8IVG
 
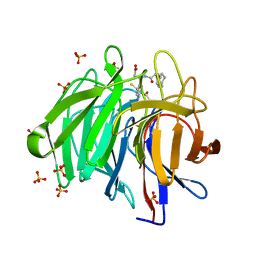 | | Methyl and Fluorine Effects in Novel Orally Bioavailable Keap1/Nrf2 PPI Inhibitor for Treatment of Chronic Kidney Disease | | Descriptor: | (3S)-3-(4-methylphenyl)-3-[2-(5,6,7,8-tetrahydronaphthalen-2-yl)ethanoylamino]propanoic acid, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Kelch-like ECH-associated protein 1, ... | | Authors: | Otake, K, Ubukata, M, Nagahashi, N, Ogawa, N, Hantani, Y, Hantani, R, Adachi, T, Nomura, A, Yamaguchi, K, Maekawa, M, Mamada, H, Motomura, T, Sato, M, Harada, K. | | Deposit date: | 2023-03-27 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Methyl and Fluorine Effects in Novel Orally Bioavailable Keap1-Nrf2 PPI Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
1BQR
 
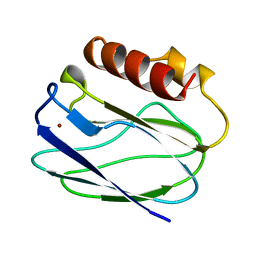 | | REDUCED PSEUDOAZURIN | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Inoue, T, Nishio, N, Hamanaka, S, Shimomura, T, Harada, S, Suzuki, S, Kohzuma, T, Shidara, S, Iwasaki, H, Kai, Y. | | Deposit date: | 1998-08-17 | | Release date: | 1999-08-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure determinations of oxidized and reduced pseudoazurins from Achromobacter cycloclastes. Concerted movement of copper site in redox forms with the rearrangement of hydrogen bond at a remote histidine.
J.Biol.Chem., 274, 1999
|
|
1BQK
 
 | | OXIDIZED PSEUDOAZURIN | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Inoue, T, Nishio, N, Hamanaka, S, Shimomura, T, Harada, S, Suzuki, S, Kohzuma, T, Shidara, S, Iwasaki, H, Kai, Y. | | Deposit date: | 1998-08-17 | | Release date: | 1999-08-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure determinations of oxidized and reduced pseudoazurins from Achromobacter cycloclastes. Concerted movement of copper site in redox forms with the rearrangement of hydrogen bond at a remote histidine.
J.Biol.Chem., 274, 1999
|
|
4YSH
 
 | |
6K3D
 
 | | Structure of multicopper oxidase mutant | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, Multicopper oxidase | | Authors: | Sakuraba, H, Ohshida, T, Satomura, T, Yoneda, K, Ohshima, T. | | Deposit date: | 2019-05-17 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Activity enhancement of multicopper oxidase from a hyperthermophile via directed evolution, and its application as the element of a high performance biocathode.
J.Biotechnol., 325, 2021
|
|
7E6Y
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 1 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E6Z
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 50 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E6X
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 4 ms structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E71
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 1 ms structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E70
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 250 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7C86
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: Dark state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RETINAL, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-05-28 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E5O
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with antibody NT-193 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NT-193 Heavy chain, NT-193 Light chain, ... | | Authors: | Kita, S, Onodera, T, Adachi, Y, Moriayma, S, Nomura, T, Tadokoro, T, Anraku, Y, Yumoto, K, Tian, C, Fukuhara, H, Suzuki, T, Tonouchi, K, Sasaki, J, Sun, L, Hashiguchi, T, Takahashi, Y, Maenaka, K. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A SARS-CoV-2 antibody broadly neutralizes SARS-related coronaviruses and variants by coordinated recognition of a virus-vulnerable site.
Immunity, 54, 2021
|
|
7WUO
 
 | |
