6QSK
 
 | |
3FVQ
 
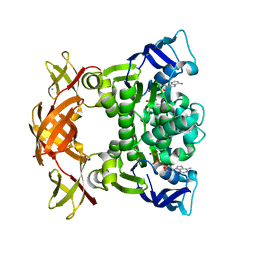 | | Crystal structure of the nucleotide binding domain FbpC complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Fe(3+) ions import ATP-binding protein fbpC | | Authors: | Newstead, S, Bilton, P, Carpenter, E.P, Campopiano, D, Iwata, S. | | Deposit date: | 2009-01-16 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into how nucleotide-binding domains power ABC transport.
Structure, 17, 2009
|
|
6ZXR
 
 | |
7OXE
 
 | |
7OYE
 
 | |
2BF6
 
 | |
5OGK
 
 | |
5OGE
 
 | |
2VK5
 
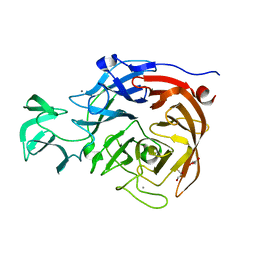 | | THE STRUCTURE OF CLOSTRIDIUM PERFRINGENS NANI SIALIDASE AND ITS CATALYTIC INTERMEDIATES | | Descriptor: | CALCIUM ION, EXO-ALPHA-SIALIDASE, GLYCEROL | | Authors: | Newstead, S.L, Potter, J.A, Wilson, J.C, Xu, G, Chien, C.H, Watts, A.G, Withers, S.G, Taylor, G.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The Structure of Clostridium Perfringens Nani Sialidase and its Catalytic Intermediates.
J.Biol.Chem., 283, 2008
|
|
2BER
 
 | | Y370G Active Site Mutant of the Sialidase from Micromonospora viridifaciens in complex with beta-Neu5Ac (sialic acid). | | Descriptor: | BACTERIAL SIALIDASE, N-acetyl-beta-neuraminic acid, SODIUM ION | | Authors: | Newstead, S, Watson, J.N, Bennet, A.J, Taylor, G.L. | | Deposit date: | 2004-11-30 | | Release date: | 2005-04-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Mechanism of Action of an Inverting Mutant Sialidase.
Biochemistry, 44, 2005
|
|
1WCQ
 
 | | Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, SIALIDASE, ... | | Authors: | Newstead, S, Watson, J.N, Bennet, A.J, Taylor, G. | | Deposit date: | 2004-11-19 | | Release date: | 2005-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Nucleophilic Mutants of the Micromonospora Viridifaciens Sialidase Operate with Retention of Configuration by Two Different Mechanisms.
Chembiochem, 6, 2005
|
|
6EI3
 
 | | Crystal structure of auto inhibited POT family peptide transporter | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Proton-dependent oligopeptide transporter family protein | | Authors: | Newstead, S, Brinth, A, Vogeley, L, Caffrey, M. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Proton movement and coupling in the POT family of peptide transporters.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2XUT
 
 | | Crystal structure of a proton dependent oligopeptide (POT) family transporter. | | Descriptor: | PROTON/PEPTIDE SYMPORTER FAMILY PROTEIN | | Authors: | Newstead, S, Drew, D, Cameron, A.D, Postis, V.L, Xia, X, Fowler, P.W, Ingram, J.C, Carpenter, E.P, Sansom, M.S.P, McPherson, M.J, Baldwin, S.A, Iwata, S. | | Deposit date: | 2010-10-21 | | Release date: | 2010-12-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal Structure of a Prokaryotic Homologue of the Mammalian Oligopeptide-Proton Symporters, Pept1 and Pept2.
Embo J., 30, 2011
|
|
1W8O
 
 | | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens | | Descriptor: | BACTERIAL SIALIDASE, CITRIC ACID, GLYCEROL, ... | | Authors: | Newstead, S, Watson, J.N, Dookhun, V, Bennet, A.J, Taylor, G. | | Deposit date: | 2004-09-24 | | Release date: | 2004-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora Viridifaciens
FEBS Lett., 577, 2004
|
|
1W8N
 
 | | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, BACTERIAL SIALIDASE, SODIUM ION, ... | | Authors: | Newstead, S, Watson, J.N, Dookhun, V, Bennet, A.J, Taylor, G. | | Deposit date: | 2004-09-24 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora Viridifaciens
FEBS Lett., 577, 2004
|
|
2BZD
 
 | |
2VK7
 
 | | THE STRUCTURE OF CLOSTRIDIUM PERFRINGENS NANI SIALIDASE AND ITS CATALYTIC INTERMEDIATES | | Descriptor: | 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CALCIUM ION, EXO-ALPHA-SIALIDASE | | Authors: | Newstead, S.L, Potter, J.A, Wilson, J.C, Xu, G, Chien, C.H, Watts, A.G, Withers, S.G, Taylor, G.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Structure of Clostridium Perfringens Nani Sialidase and its Catalytic Intermediates.
J.Biol.Chem., 283, 2008
|
|
2VK6
 
 | | THE STRUCTURE OF CLOSTRIDIUM PERFRINGENS NANI SIALIDASE AND ITS CATALYTIC INTERMEDIATES | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, EXO-ALPHA-SIALIDASE, ... | | Authors: | Newstead, S.L, Potter, J.A, Wilson, J.C, Xu, G, Chien, C.H, Watts, A.G, Withers, S.G, Taylor, G.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of Clostridium Perfringens Nani Sialidase and its Catalytic Intermediates.
J.Biol.Chem., 283, 2008
|
|
4UVM
 
 | | In meso crystal structure of the POT family transporter PepTSo | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, GLUTATHIONE UPTAKE TRANSPORTER | | Authors: | Lyons, J.A, Solcan, N, Caffrey, M, Newstead, S. | | Deposit date: | 2014-08-07 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Gating Topology of the Proton-Coupled Oligopeptide Symporters.
Structure, 23, 2015
|
|
6BQO
 
 | | Structure of a dual topology fluoride channel with monobody S8 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, FLUORIDE ION, Fluoride ion transporter CrcB, ... | | Authors: | Stockbridge, R.B, Newstead, S, McIlwain, B.C. | | Deposit date: | 2017-11-28 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cork-in-Bottle Occlusion of Fluoride Ion Channels by Crystallization Chaperones.
Structure, 26, 2018
|
|
5L7I
 
 | | Structure of human Smoothened in complex with Vismodegib | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloranyl-~{N}-(4-chloranyl-3-pyridin-2-yl-phenyl)-4-methylsulfonyl-benzamide, SODIUM ION, ... | | Authors: | Byrne, E.X.B, Sircar, R, Miller, P.S, Hedger, G, Luchetti, G, Nachtergaele, S, Tully, M.D, Mydock-McGrane, L, Covey, D.F, Rambo, R.F, Sansom, M.S.P, Newstead, S, Rohatgi, R, Siebold, C. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of Smoothened regulation by its extracellular domains.
Nature, 535, 2016
|
|
5L7D
 
 | | Structure of human Smoothened in complex with cholesterol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, SODIUM ION, ... | | Authors: | Byrne, E.F.X, Sircar, R, Miller, P.S, Hedger, G, Luchetti, G, Nachtergaele, S, Tully, M.D, Mydock-McGrane, L, Covey, D.F, Rambo, R.P, Sansom, M.S.P, Newstead, S, Rohatgi, R, Siebold, C. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of Smoothened regulation by its extracellular domains.
Nature, 535, 2016
|
|
8OMU
 
 | |
8P6A
 
 | |
6Y7V
 
 | | Crystal structure of the KDEL receptor bound to HDEL peptide at pH 6.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON DIOXIDE, ER lumen protein-retaining receptor 2, ... | | Authors: | Braeuer, P, Newstead, S. | | Deposit date: | 2020-03-02 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | A signal capture and proofreading mechanism for the KDEL-receptor explains selectivity and dynamic range in ER retrieval.
Elife, 10, 2021
|
|
