7DIH
 
 | |
5YGQ
 
 | |
7F30
 
 | | Crystal structure of OxdB E85A in complex with Z-2- (3-bromophenyl) propanal oxime | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phenylacetaldoxime dehydratase, Z-2-(3-bromophenyl) propanal oxime | | Authors: | Muraki, N, Matsui, D, Asano, Y, Aono, S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structural analysis of aldoxime dehydratase from Bacillus sp. OxB-1: Importance of surface residues in optimization for crystallization.
J.Inorg.Biochem., 230, 2022
|
|
7F2Y
 
 | | Crystal structure of OxdB E85A mutant (form I) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phenylacetaldoxime dehydratase | | Authors: | Muraki, N, Matsui, D, Asano, Y, Aono, S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structural analysis of aldoxime dehydratase from Bacillus sp. OxB-1: Importance of surface residues in optimization for crystallization.
J.Inorg.Biochem., 230, 2022
|
|
7F2Z
 
 | | Crystal structure of OxdB E85A mutant (form II) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phenylacetaldoxime dehydratase | | Authors: | Muraki, N, Matsui, D, Asano, Y, Aono, S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structural analysis of aldoxime dehydratase from Bacillus sp. OxB-1: Importance of surface residues in optimization for crystallization.
J.Inorg.Biochem., 230, 2022
|
|
7DVS
 
 | | Crystal structure of Apo (heme-free) PefR | | Descriptor: | MarR family transcriptional regulator | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival.
Commun Biol, 4, 2021
|
|
3AEQ
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N, ... | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AES
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AEK
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N, ... | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AET
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AEU
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AER
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AB1
 
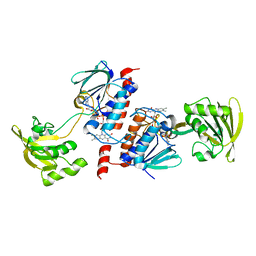 | | Crystal Structure of Ferredoxin NADP+ Oxidoreductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase | | Authors: | Muraki, N, Seo, D, Kurisu, G. | | Deposit date: | 2009-11-30 | | Release date: | 2010-11-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Asymmetric dimeric structure of ferredoxin-NAD(P)+ oxidoreductase from the green sulfur bacterium Chlorobaculum tepidum: implications for binding ferredoxin and NADP+
J.Mol.Biol., 401, 2010
|
|
5AZ3
 
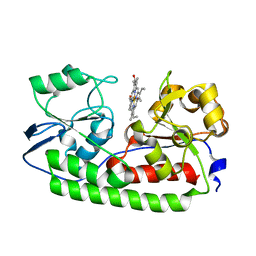 | | Crystal structure of heme binding protein HmuT | | Descriptor: | ABC-type transporter, periplasmic component, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2015-09-18 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | Structural Basis for Heme Recognition by HmuT Responsible for Heme Transport to the Heme Transporter in Corynebacterium glutamicum
Chem Lett., 2015
|
|
5B51
 
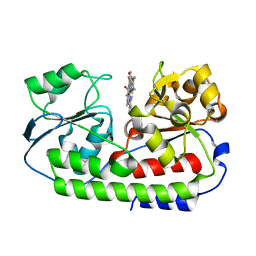 | | Crystal structure of heme binding protein HmuT R242A mutant | | Descriptor: | ABC-type transporter, periplasmic component, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Characterization of Heme Environmental Mutants of CgHmuT that Shuttles Heme Molecules to Heme Transporters
Int J Mol Sci, 17, 2016
|
|
5B4Z
 
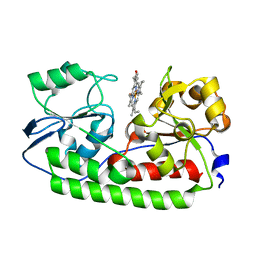 | | Crystal structure of heme binding protein HmuT H141A mutant | | Descriptor: | ABC-type transporter, periplasmic component, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Characterization of Heme Environmental Mutants of CgHmuT that Shuttles Heme Molecules to Heme Transporters
Int J Mol Sci, 17, 2016
|
|
5B50
 
 | | Crystal structure of heme binding protein HmuT Y240A | | Descriptor: | ABC-type transporter, periplasmic component, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterization of Heme Environmental Mutants of CgHmuT that Shuttles Heme Molecules to Heme Transporters
Int J Mol Sci, 17, 2016
|
|
3VO2
 
 | |
3VO1
 
 | |
6J1G
 
 | |
6J1F
 
 | | Crystal structure of HypX from Aquifex aeolicus in complex with Tetrahydrofolic acid | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, COENZYME A, GLYCEROL, ... | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of HypX responsible for CO biosynthesis in the maturation of NiFe-hydrogenase.
Commun Biol, 2, 2019
|
|
6J1I
 
 | |
6J1J
 
 | | Crystal structure of HypX from Aquifex aeolicus, A392F-I419F variant in complex with Tetrahydrofolic acid | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, COENZYME A, GLYCEROL, ... | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of HypX responsible for CO biosynthesis in the maturation of NiFe-hydrogenase.
Commun Biol, 2, 2019
|
|
6J1H
 
 | |
6J0P
 
 | |
