4ZY1
 
 | |
4ZX3
 
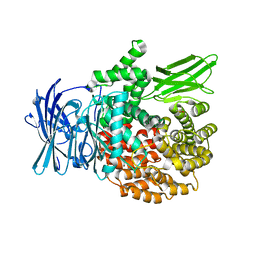 | |
5CDZ
 
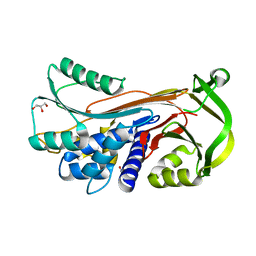 | | Crystal structure of conserpin in the latent state | | Descriptor: | Conserpin in the latent state, GLYCEROL | | Authors: | Porebski, B.T, McGowan, S, Keleher, S, Buckle, A.M. | | Deposit date: | 2015-07-06 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Smoothing a rugged protein folding landscape by sequence-based redesign.
Sci Rep, 6, 2016
|
|
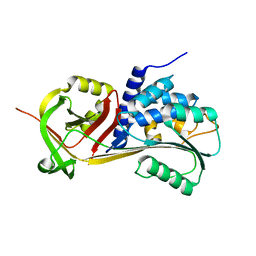
5CE0
 
 | |
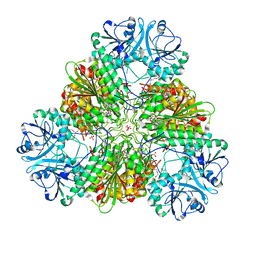
5CBM
 
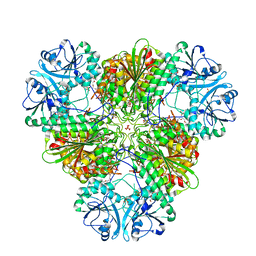 | | Crystal structure of PfA-M17 with virtual ligand inhibitor | | Descriptor: | (2S)-2-{[(R)-[(R)-amino(phenyl)methyl](hydroxy)phosphoryl]methyl}-4-methylpentanoic acid, CARBONATE ION, GLYCEROL, ... | | Authors: | Ruggeri, C, Drinkwater, N, McGowan, S. | | Deposit date: | 2015-07-01 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and Validation of a Potent Dual Inhibitor of the P. falciparum M1 and M17 Aminopeptidases Using Virtual Screening.
Plos One, 10, 2015
|
|
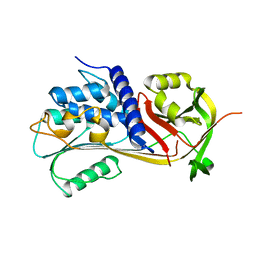
5CDX
 
 | |
4ZQT
 
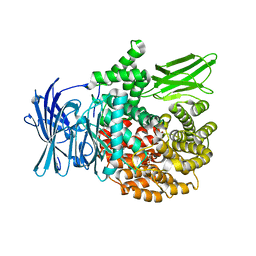 | | Crystal structure of PfA-M1 with virtual ligand inhibitor | | Descriptor: | (2R)-2-{[(R)-[(R)-amino(phenyl)methyl](hydroxy)phosphoryl]methyl}-4-methylpentanoic acid, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Ruggeri, C, Drinkwater, N, McGowan, S. | | Deposit date: | 2015-05-11 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Identification and Validation of a Potent Dual Inhibitor of the P. falciparum M1 and M17 Aminopeptidases Using Virtual Screening.
Plos One, 10, 2015
|
|
