1COH
 
 | |
1NIH
 
 | |
1CHC
 
 | |
1IBE
 
 | |
1LR0
 
 | | Pseudomonas aeruginosa TolA Domain III, Seleno-methionine Derivative | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TolA protein, ZINC ION | | Authors: | Witty, M, Sanz, C, Shah, A, Grossman, J.G, Mizuguchi, K, Perham, R.N, Luisi, B. | | Deposit date: | 2002-05-14 | | Release date: | 2002-05-29 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Structure of the periplasmic domain of Pseudomonas aeruginosa TolA: evidence for an evolutionary relationship with the TonB transporter protein.
EMBO J., 21, 2002
|
|
1EK9
 
 | | 2.1A X-RAY STRUCTURE OF TOLC: AN INTEGRAL OUTER MEMBRANE PROTEIN AND EFFLUX PUMP COMPONENT FROM ESCHERICHIA COLI | | Descriptor: | OUTER MEMBRANE PROTEIN TOLC | | Authors: | Koronakis, V, Sharff, A.J, Koronakis, E, Luisi, B, Hughes, C. | | Deposit date: | 2000-03-07 | | Release date: | 2000-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export.
Nature, 405, 2000
|
|
6TNN
 
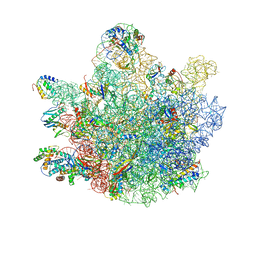 | | Mini-RNase III (Mini-III) bound to 50S ribosome with precursor 23S rRNA | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Oerum, S, Dendooven, T, Gilet, L, Catala, M, Degut, C, Trinquier, A, Barraud, P, Luisi, B, Condon, C, Tisne, C. | | Deposit date: | 2019-12-09 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
6TPQ
 
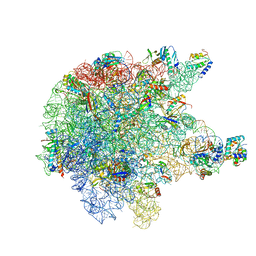 | | RNase M5 bound to 50S ribosome with precursor 5S rRNA | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Oerum, S, Dendooven, T, Gilet, L, Catala, M, Degut, C, Trinquier, A, Barraud, P, Luisi, B, Condon, C, Tisne, C. | | Deposit date: | 2019-12-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
4OWG
 
 | |
2VDE
 
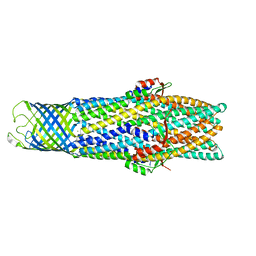 | | Crystal Structure of the Open State of TolC Outer Membrane Component of Mutlidrug Efflux Pumps | | Descriptor: | CHLORIDE ION, OUTER MEMBRANE PROTEIN TOLC | | Authors: | Bavro, V.N, Pietras, Z, Furnham, N, Perez-Cano, L, Fernandez-Recio, J, Pei, X.Y, Truer, R, Misra, R, Luisi, B. | | Deposit date: | 2007-10-04 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Assembly and Channel Opening in a Bacterial Drug Efflux Machine.
Mol.Cell, 30, 2008
|
|
2VDD
 
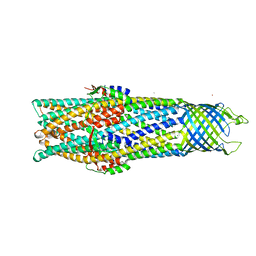 | | Crystal Structure of the Open State of TolC Outer Membrane Component of Mutlidrug Efflux Pumps | | Descriptor: | CHLORIDE ION, OUTER MEMBRANE PROTEIN TOLC, POTASSIUM ION | | Authors: | Bavro, V.N, Pietras, Z, Furnham, N, Perez-Cano, L, Fernandez-Recio, J, Pei, X.Y, Truer, R, Misra, R, Luisi, B. | | Deposit date: | 2007-10-04 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Assembly and Channel Opening in a Bacterial Drug Efflux Machine.
Mol.Cell, 30, 2008
|
|
1W08
 
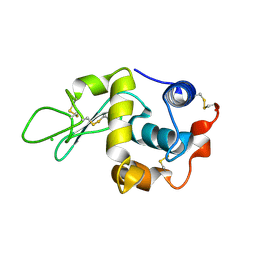 | | STRUCTURE OF T70N HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME | | Authors: | Johnson, R, Christodoulou, J, Luisi, B, Dumoulin, M, Caddy, G, Alcocer, M, Murtagh, G, Archer, D.B, Dobson, C.M. | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rationalising Lysozyme Amyloidosis: Insights from the Structure and Solution Dynamics of T70N Lysozyme.
J.Mol.Biol., 352, 2005
|
|
1E9I
 
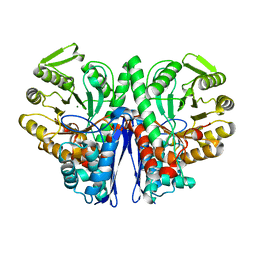 | | Enolase from E.coli | | Descriptor: | ENOLASE, MAGNESIUM ION, SULFATE ION | | Authors: | Kuhnel, K, Carpousis, A.J, Luisi, B. | | Deposit date: | 2000-10-17 | | Release date: | 2001-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure of the Escherichia Coli RNA Degradosome Component Enolase
J.Mol.Biol., 313, 2001
|
|
352D
 
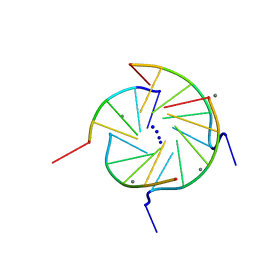 | | THE CRYSTAL STRUCTURE OF A PARALLEL-STRANDED PARALLEL-STRANDED GUANINE TETRAPLEX AT 0.95 ANGSTROM RESOLUTION | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3'), SODIUM ION | | Authors: | Phillips, K, Dauter, Z, Murchie, A.I.H, Lilley, D.M.J, Luisi, B. | | Deposit date: | 1997-09-04 | | Release date: | 1997-11-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The crystal structure of a parallel-stranded guanine tetraplex at 0.95 A resolution.
J.Mol.Biol., 273, 1997
|
|
244D
 
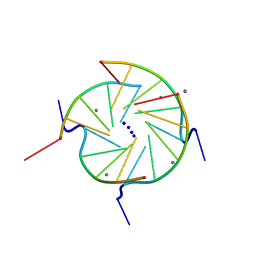 | | THE HIGH-RESOLUTION CRYSTAL STRUCTURE OF A PARALLEL-STRANDED GUANINE TETRAPLEX | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3'), SODIUM ION | | Authors: | Laughlan, G, Murchie, A.I.H, Norman, D.G, Moore, M.H, Moody, P.C.E, Lilley, D.M.J, Luisi, B. | | Deposit date: | 1995-10-19 | | Release date: | 1996-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The high-resolution crystal structure of a parallel-stranded guanine tetraplex.
Science, 265, 1994
|
|
5NC5
 
 | |
5NB9
 
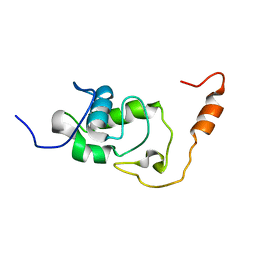 | | Structure of the N-terminal domain of the Escherichia Coli ProQ RNA binding protein | | Descriptor: | RNA chaperone ProQ | | Authors: | Gonzales, G, Hardwick, S, Maslen, S, Skehel, M, Holmqvist, E, Vogel, J, Bateman, A, Luisi, B, Broadhurst, R. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the Escherichia coli ProQ RNA-binding protein.
RNA, 23, 2017
|
|
5NBB
 
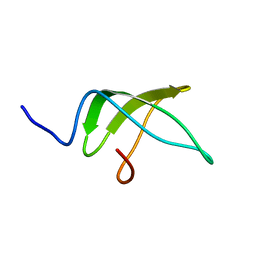 | | Structure of the C-terminal domain of the Escherichia Coli ProQ RNA binding protein | | Descriptor: | RNA chaperone ProQ | | Authors: | Gonzales, G, Hardwick, S, Maslen, S, Skehel, M, Holmqvist, E, Vogel, J, Bateman, A, Luisi, B, Broadhurst, R. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the Escherichia coli ProQ RNA-binding protein.
RNA, 23, 2017
|
|
