4WXO
 
 | |
4WXW
 
 | |
7JRE
 
 | |
7LW2
 
 | |
7MG0
 
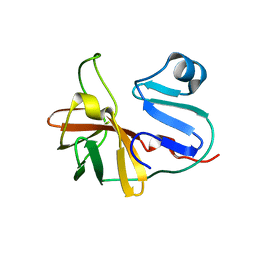 | |
6PQR
 
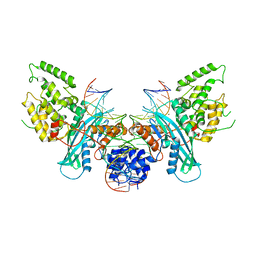 | | Cryo-EM structure of HzTransib/intact TIR substrate DNA pre-reaction complex (PRC) | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*AP*TP*CP*TP*CP*AP*CP*GP*GP*TP*GP*GP*AP*TP*CP*GP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*CP*GP*AP*TP*CP*CP*AP*CP*CP*GP*TP*GP*AP*GP*AP*TP*CP*TP*AP*G)-3'), DNA-mediated transposase, ... | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
2ZY1
 
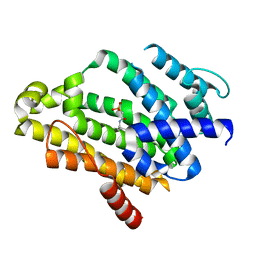 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with bisphosphonate BPH-830 | | Descriptor: | Dehydrosqualene synthase, dipotassium (2-oxo-2-{[3-(3-phenoxyphenyl)propyl]amino}ethyl)phosphonate | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2009-01-10 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Inhibition of staphyloxanthin virulence factor biosynthesis in Staphylococcus aureus: in vitro, in vivo, and crystallographic results.
J.Med.Chem., 52, 2009
|
|
3ADZ
 
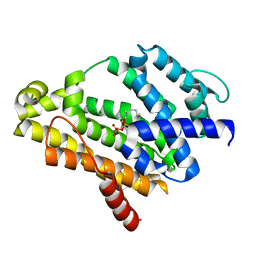 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with intermediate PSPP | | Descriptor: | Dehydrosqualene synthase, MAGNESIUM ION, {(1R,2R,3R)-2-[(3E)-4,8-dimethylnona-3,7-dien-1-yl]-2-methyl-3-[(1E,5E)-2,6,10-trimethylundeca-1,5,9-trien-1-yl]cyclopropyl}methyl trihydrogen diphosphate | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2010-01-31 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3AE0
 
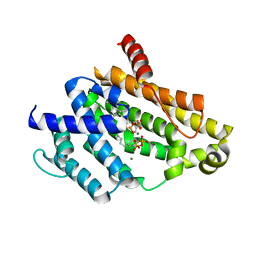 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with geranylgeranyl thiopyrophosphate | | Descriptor: | Dehydrosqualene synthase, MAGNESIUM ION, phosphonooxy-[(10E)-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraenyl]sulfanyl-phosphinic acid | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2010-01-31 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ACW
 
 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with BPH-651 | | Descriptor: | (3R)-3-biphenyl-4-yl-1-azabicyclo[2.2.2]octan-3-ol, Dehydrosqualene synthase | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2010-01-13 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ACX
 
 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with BPH-673 | | Descriptor: | Dehydrosqualene synthase, N-(1-methylethyl)-3-[(3-prop-2-en-1-ylbiphenyl-4-yl)oxy]propan-1-amine | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2010-01-13 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ACY
 
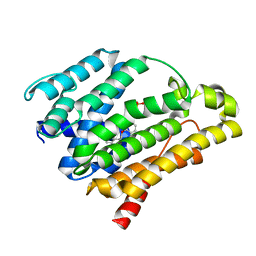 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with BPH-702 | | Descriptor: | (1R)-4-[3-(2-benzylphenoxy)phenyl]-1-phosphonobutane-1-sulfonic acid, Dehydrosqualene synthase, MAGNESIUM ION | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2010-01-13 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6PQY
 
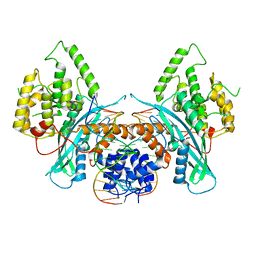 | | Cryo-EM structure of HzTransib/TIR DNA transposon end complex (TEC) | | Descriptor: | DNA (5'-D(P*CP*AP*CP*GP*GP*TP*GP*GP*AP*TP*CP*GP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*CP*GP*AP*TP*CP*CP*AP*CP*CP*GP*TP*G)-3'), Putative DNA-mediated transposase | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
6PR5
 
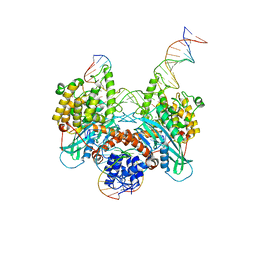 | | Cryo-EM structure of HzTransib strand transfer complex (STC) | | Descriptor: | DNA (30-MER), DNA (39-MER), DNA (5'-D(*GP*AP*TP*CP*TP*GP*GP*CP*CP*TP*AP*GP*AP*TP*CP*TP*CP*A)-3'), ... | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
6PQU
 
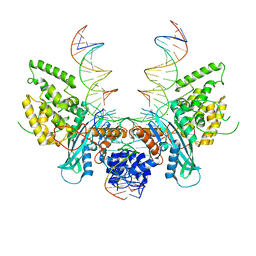 | | Cryo-EM structure of HzTransib/nicked TIR substrate DNA pre-reaction complex (PRC) | | Descriptor: | DNA (5'-D(P*AP*TP*CP*TP*GP*GP*CP*CP*TP*AP*GP*AP*TP*CP*T)-3'), DNA (5'-D(P*CP*AP*CP*GP*GP*TP*GP*GP*AP*TP*CP*GP*AP*AP*AP*A)-3'), DNA-mediated transposase, ... | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
6PQX
 
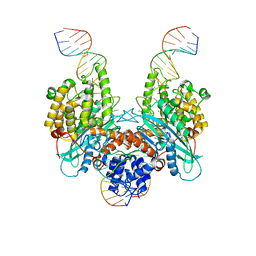 | | Cryo-EM structure of HzTransib/nicked TIR substrate DNA hairpin forming complex (HFC) | | Descriptor: | CALCIUM ION, DNA (5'-D(P*CP*AP*CP*GP*GP*TP*GP*GP*AP*TP*CP*GP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*CP*TP*GP*GP*CP*CP*TP*AP*GP*AP*TP*CP*T)-3'), ... | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
6ILX
 
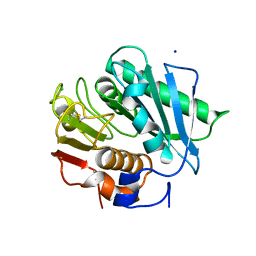 | | Crystal structure of PETase W159F mutant from Ideonella sakaiensis | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SODIUM ION | | Authors: | Liu, C.C, Shi, C. | | Deposit date: | 2018-10-20 | | Release date: | 2019-03-27 | | Last modified: | 2020-09-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional characterization of polyethylene terephthalate hydrolase from Ideonella sakaiensis.
Biochem. Biophys. Res. Commun., 508, 2019
|
|
6PQN
 
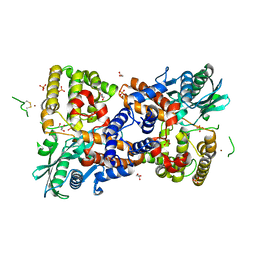 | | Crystal structure of HzTransib transposase | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative DNA-mediated transposase, ... | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-09 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
7N0D
 
 | |
2ZCS
 
 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with bisphosphonate BPH-700 | | Descriptor: | Dehydrosqualene synthase, tripotassium (1R)-4-biphenyl-4-yl-1-phosphonatobutane-1-sulfonate | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H, Oldfield, E. | | Deposit date: | 2007-11-11 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence.
Science, 319, 2008
|
|
2ZCQ
 
 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with bisphosphonate BPH-652 | | Descriptor: | (1R)-4-(3-phenoxyphenyl)-1-phosphonobutane-1-sulfonic acid, Dehydrosqualene synthase, MAGNESIUM ION | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H, Oldfield, E. | | Deposit date: | 2007-11-11 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence.
Science, 319, 2008
|
|
2ZCR
 
 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with bisphosphonate BPH-698 | | Descriptor: | Dehydrosqualene synthase, MAGNESIUM ION, tripotassium (1R)-4-(4'-butylbiphenyl-4-yl)-1-phosphonatobutane-1-sulfonate | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H, Oldfield, E. | | Deposit date: | 2007-11-11 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence.
Science, 319, 2008
|
|
8DZJ
 
 | |
3VJE
 
 | | Crystal structure of the Y248A mutant of C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus in complex with zaragozic acid A | | Descriptor: | Dehydrosqualene synthase, Zaragozic acid A | | Authors: | Liu, C.I, Jeng, W.Y, Chang, W.J, Wang, A.H.J. | | Deposit date: | 2011-10-14 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Binding modes of zaragozic acid A to human squalene synthase and staphylococcal dehydrosqualene synthase
J.Biol.Chem., 287, 2012
|
|
3VJD
 
 | | Crystal structure of the Y248A mutant of C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus | | Descriptor: | Dehydrosqualene synthase, L(+)-TARTARIC ACID | | Authors: | Liu, C.I, Jeng, W.Y, Chang, W.J, Wang, A.H.J. | | Deposit date: | 2011-10-14 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Binding modes of zaragozic acid A to human squalene synthase and staphylococcal dehydrosqualene synthase
J.Biol.Chem., 287, 2012
|
|
