4RY2
 
 | |
4S0F
 
 | |
4W9A
 
 | | Crystal structure of Gamma-B Crystallin expressed in E. coli based on mRNA variant 2 | | Descriptor: | Gamma-crystallin B | | Authors: | Kudlinzki, D, Buhr, F, Linhard, V.L, Jha, S, Komar, A.A, Schwalbe, H. | | Deposit date: | 2014-08-27 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Two synonymous gene variants encode proteins with identical sequence, but different folding conformations.
To Be Published
|
|
8FD9
 
 | |
8FF0
 
 | |
3BIK
 
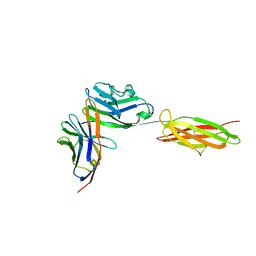 | | Crystal Structure of the PD-1/PD-L1 Complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BIS
 
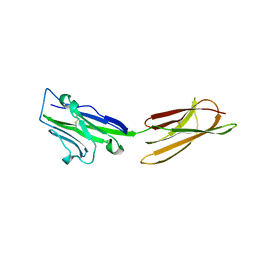 | | Crystal Structure of the PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5FD6
 
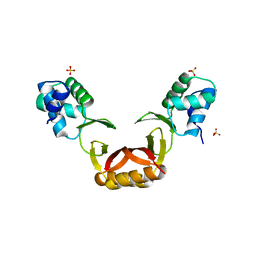 | |
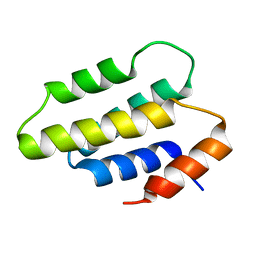
8QRY
 
 | |
6PCK
 
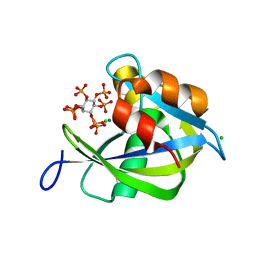 | | Crystal structure of human diphosphoinositol polyphosphate phosphohydrolase 1 in complex with 1-IP7 | | Descriptor: | (1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Dollins, D.E, Neubauer, J, Dong, J, York, J.D. | | Deposit date: | 2019-06-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Vip1 is a kinase and pyrophosphatase switch that regulates inositol diphosphate signaling.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PCL
 
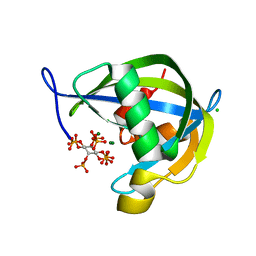 | | Crystal structure of human diphosphoinositol polyphosphate phosphohydrolase 1 in complex with 5-IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Dollins, D.E, Neubauer, J, Dong, J, York, J.D. | | Deposit date: | 2019-06-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Vip1 is a kinase and pyrophosphatase switch that regulates inositol diphosphate signaling.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5HB8
 
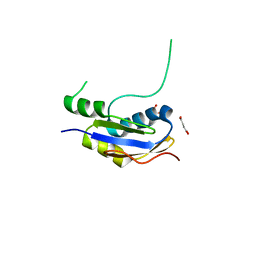 | |
5HB0
 
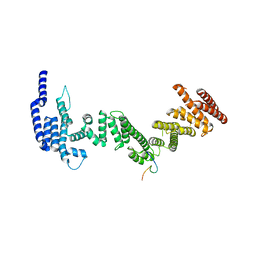 | |
5HAX
 
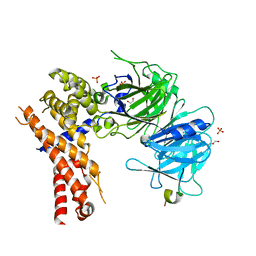 | | Crystal structure of Chaetomium thermophilum Nup170 NTD-Nup53 complex | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleoporin NUP170, ... | | Authors: | Lin, D.H, Mobbs, G, Hoelz, A. | | Deposit date: | 2015-12-31 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Architecture of the symmetric core of the nuclear pore.
Science, 352, 2016
|
|
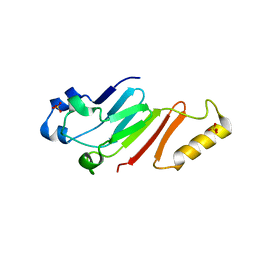
5HB6
 
 | |
5HAY
 
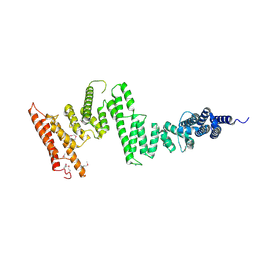 | |
5HB3
 
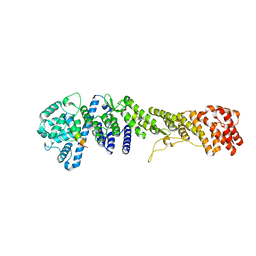 | |
5HB7
 
 | |
5HAZ
 
 | | Structure of Chaetomium thermophilum Nup170 CTD | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nucleoporin NUP170, ... | | Authors: | Lin, D.H, Hoelz, A. | | Deposit date: | 2015-12-31 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Architecture of the symmetric core of the nuclear pore.
Science, 352, 2016
|
|
5HB5
 
 | |
5HB2
 
 | |
5HB1
 
 | |
7B67
 
 | | Structure of NUDT15 V18_V19insGV Mutant in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, MAGNESIUM ION, Nucleotide triphosphate diphosphatase NUDT15, ... | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7B65
 
 | | Structure of NUDT15 R139C Mutant in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, Nucleotide triphosphate diphosphatase NUDT15 | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7B63
 
 | | Structure of NUDT15 in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, MAGNESIUM ION, Probable 8-oxo-dGTP diphosphatase NUDT15 | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
