7Z88
 
 | | DNA-PK in the intermediate state | | Descriptor: | (~{S})-[2-chloranyl-4-fluoranyl-5-(7-morpholin-4-ylquinazolin-4-yl)phenyl]-(6-methoxypyridazin-3-yl)methanol, DNA (26-MER), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Liang, S, Blundell, T.L. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Human DNA-dependent protein kinase activation mechanism.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z87
 
 | | DNA-PK in the active state | | Descriptor: | (~{S})-[2-chloranyl-4-fluoranyl-5-(7-morpholin-4-ylquinazolin-4-yl)phenyl]-(6-methoxypyridazin-3-yl)methanol, DNA (26-MER), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Liang, S, Blundell, T.L. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Human DNA-dependent protein kinase activation mechanism.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7OTP
 
 | | DNA-PKcs in complex with ATPgammaS-Mg | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTY
 
 | | DNA-PKcs in complex with M3814 | | Descriptor: | (~{S})-[2-chloranyl-4-fluoranyl-5-(7-morpholin-4-ylquinazolin-4-yl)phenyl]-(6-methoxypyridazin-3-yl)methanol, DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTM
 
 | | Cryo-EM structure of DNA-PKcs in complex with NU7441 | | Descriptor: | 8-(dibenzo[b,d]thiophen-4-yl)-2-(morpholin-4-yl)-4H-chromen-4-one, DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTV
 
 | | DNA-PKcs in complex with wortmannin | | Descriptor: | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4, 3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE, DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTW
 
 | | DNA-PKcs in complex with AZD7648 | | Descriptor: | 7-methyl-2-[(7-methyl-[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(oxan-4-yl)purin-8-one, DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7RSX
 
 | | HIV-1 gp120 complex with CJF-III-049-S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN GP120, N~1~-{(1R,2R,3S)-2-(carbamimidamidomethyl)-3-[(3S)-3,4-dihydroxybutyl]-5-[(methylamino)methyl]-2,3-dihydro-1H-inden-1-yl}-N~2~-(4-chloro-3-fluorophenyl)ethanediamide | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of gp120 Residue His105 as a Novel Target for HIV-1 Neutralization by Small-Molecule CD4-Mimics.
Acs Med.Chem.Lett., 12, 2021
|
|
7RSZ
 
 | | HIV-1 gp120 complex with CJF-II-204 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 gp120 Clade C1086, N~1~-{(1R,2R,3S)-2-(carbamimidamidomethyl)-3-[(3R)-3,4-dihydroxybutyl]-5-[(methylamino)methyl]-2,3-dihydro-1H-inden-1-yl}-N~2~-(4-chloro-3-fluorophenyl)ethanediamide, ... | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Identification of gp120 Residue His105 as a Novel Target for HIV-1 Neutralization by Small-Molecule CD4-Mimics.
Acs Med.Chem.Lett., 12, 2021
|
|
7RSY
 
 | | HIV-1 gp120 complex with CJF-III-049-R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 gp120 Clade C1086, N~1~-{(1R,2R,3S)-2-(carbamimidamidomethyl)-3-[(3R)-3,4-dihydroxybutyl]-5-[(methylamino)methyl]-2,3-dihydro-1H-inden-1-yl}-N~2~-(4-chloro-3-fluorophenyl)ethanediamide | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of gp120 Residue His105 as a Novel Target for HIV-1 Neutralization by Small-Molecule CD4-Mimics.
Acs Med.Chem.Lett., 12, 2021
|
|
5ARL
 
 | |
3W2D
 
 | | Crystal Structure of Staphylococcal Eenterotoxin B in complex with a novel neutralization monoclonal antibody Fab fragment | | Descriptor: | Enterotoxin type B, Monoclonal Antibody 3E2 Fab figment heavy chain, Monoclonal Antibody 3E2 Fab figment light chain, ... | | Authors: | Liang, S.Y, Hu, S, Dai, J.X, Guo, Y.J, Lou, Z.Y. | | Deposit date: | 2012-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2014-04-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the neutralization and specificity of Staphylococcal enterotoxin B against its MHC Class II binding site.
MAbs, 6, 2014
|
|
4G3Y
 
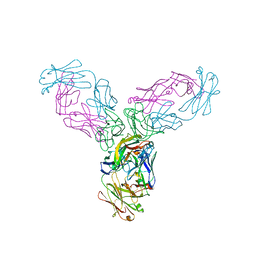 | | Crystal structure of TNF-alpha in complex with Infliximab Fab fragment | | Descriptor: | Tumor necrosis factor, infliximab Fab H, infliximab Fab L | | Authors: | Liang, S.Y, Dai, J.X, Guo, Y.J, Lou, Z.Y. | | Deposit date: | 2012-07-15 | | Release date: | 2013-03-27 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for treating tumor necrosis factor alpha (TNFalpha)-associated diseases with the therapeutic antibody infliximab
J.Biol.Chem., 288, 2013
|
|
5F4R
 
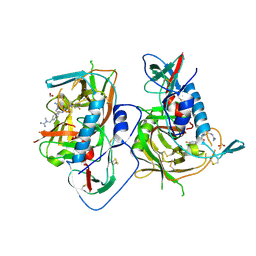 | | HIV-1 gp120 complex with BNW-IV-147 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN GP120 of HIV-1 clade C, FORMIC ACID, ... | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Small-Molecule CD4-Mimics: Structure-Based Optimization of HIV-1 Entry Inhibition.
ACS Med Chem Lett, 7, 2016
|
|
5F4U
 
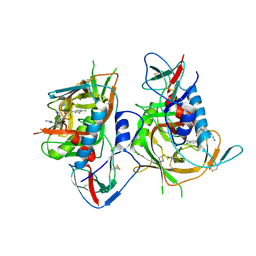 | | HIV-1 gp120 complex with BNM-IV-197 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN GP120 of HIV-1 clade C, ~{N}'-[(1~{R},2~{R})-2-(carbamimidamidomethyl)-6-[[carbamimidoyl(methyl)amino]methyl]-2,3-dihydro-1~{H}-inden-1-yl]-~{N}-(4-chloranyl-3-fluoranyl-phenyl)ethanediamide | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Small-Molecule CD4-Mimics: Structure-Based Optimization of HIV-1 Entry Inhibition.
ACS Med Chem Lett, 7, 2016
|
|
5F4P
 
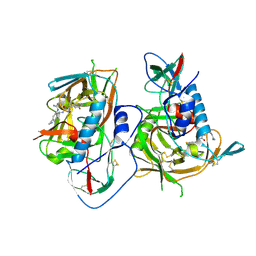 | | HIV-1 gp120 complex with BNM-III-170 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN GP120 of HIV-1 clade C, ~{N}'-[(1~{R},2~{R})-2-(carbamimidamidomethyl)-5-(methylaminomethyl)-2,3-dihydro-1~{H}-inden-1-yl]-~{N}-(4-chloranyl-3-fluoranyl-phenyl)ethanediamide | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Small-Molecule CD4-Mimics: Structure-Based Optimization of HIV-1 Entry Inhibition.
Acs Med.Chem.Lett., 7, 2016
|
|
5F4L
 
 | | HIV-1 gp120 complex with JP-III-048 | | Descriptor: | ENVELOPE GLYCOPROTEIN GP120 of HIV-1 clade C, ~{N}'-[(1~{R},2~{R})-2-(carbamimidamidomethyl)-6-(methylaminomethyl)-2,3-dihydro-1~{H}-inden-1-yl]-~{N}-(4-chloranyl-3-fluoranyl-phenyl)ethanediamide | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-30 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Small-Molecule CD4-Mimics: Structure-Based Optimization of HIV-1 Entry Inhibition.
Acs Med.Chem.Lett., 7, 2016
|
|
8KEI
 
 | | Cryo-EM structure of NADPH oxidase 2 in complex with p22phox and EROS | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liang, S.Y, Liu, A.J, Liu, Y.Z, Ye, R.D. | | Deposit date: | 2023-08-11 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM structure of NADPH oxidase 2 in complex with p22phox and EROS
To Be Published
|
|
5AR6
 
 | | crystal structure of porcine RNase 4 | | Descriptor: | 1,2-ETHANEDIOL, RIBONUCLEASE 4 | | Authors: | Liang, S, Acharya, K.R. | | Deposit date: | 2015-09-24 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis of Substrate Specificity in Porcine Rnase 4.
FEBS J., 283, 2016
|
|
5ARK
 
 | |
5ARJ
 
 | |
4FP5
 
 | | Heat-labile enterotoxin ILT-IIbB5 S74A mutant | | Descriptor: | Heat-labile enterotoxin IIB, B chain, SULFATE ION | | Authors: | Cody, V, Pace, J, Nawar, H, Liang, S, Connell, T, Hajishengallis, G. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-activity correlations of variant forms of the B pentamer of Escherichia coli type II heat-labile enterotoxin LT-IIb with Toll-like receptor 2 binding.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7P0K
 
 | | Crystal structure of Autotaxin (ENPP2) with 18F-labeled positron emission tomography ligand | | Descriptor: | 2-[[2-ethyl-6-[4-[2-[(3~{R})-3-fluoranylpyrrolidin-1-yl]-2-oxidanylidene-ethyl]piperazin-1-yl]imidazo[1,2-a]pyridin-3-yl]-methyl-amino]-4-(4-fluorophenyl)-2,3-dihydro-1,3-thiazole-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salgado-Polo, F, Shao, T, Xiao, Z, Van, R, Chen, J, Rong, J, Haider, A, Shao, Y, Josephson, L, Perrakis, A, Liang, S.H. | | Deposit date: | 2021-06-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Imaging Autotaxin In Vivo with 18 F-Labeled Positron Emission Tomography Ligands
J Med Chem, 64, 2021
|
|
1I25
 
 | |
1RYG
 
 | |
