7XST
 
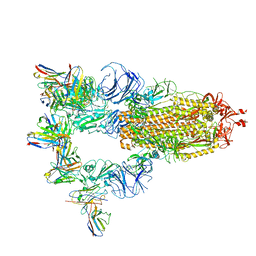 | |
7XMX
 
 | | Cryo-EM structure of SARS-CoV-2 spike glycoprotein in complex with three F61 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F61 heavy chain, F61 light chain, ... | | Authors: | Wang, X, Li, X. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages.
Cell Discov, 8, 2022
|
|
7XMZ
 
 | | Cryo-EM structure of SARS-CoV-2 spike glycoprotein in complex with three D2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D2 heavy chain, D2 light chain, ... | | Authors: | Wang, X, Li, X. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages.
Cell Discov, 8, 2022
|
|
4EDL
 
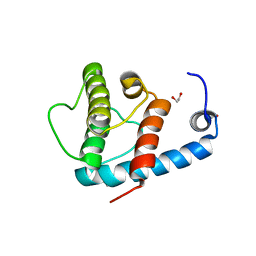 | | Crystal structure of beta-parvin CH2 domain | | Descriptor: | 1,2-ETHANEDIOL, Beta-parvin | | Authors: | Stiegler, A.L, Draheim, K.M, Li, X, Chayen, N.E, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2012-03-27 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for paxillin binding and focal adhesion targeting of beta-parvin.
J.Biol.Chem., 287, 2012
|
|
4EDN
 
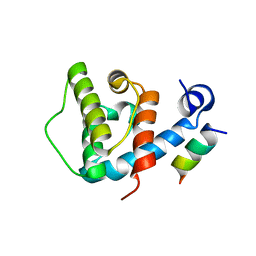 | | Crystal structure of beta-parvin CH2 domain in complex with paxillin LD1 motif | | Descriptor: | Beta-parvin, Paxillin, SULFATE ION | | Authors: | Stiegler, A.L, Draheim, K.M, Li, X, Chayen, N.E, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2012-03-27 | | Release date: | 2012-08-08 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for paxillin binding and focal adhesion targeting of beta-parvin.
J.Biol.Chem., 287, 2012
|
|
4PIR
 
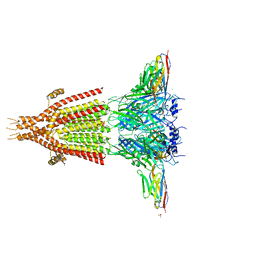 | | X-ray structure of the mouse serotonin 5-HT3 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Hassaine, G, Deluz, C, Grasso, L, Wyss, R, Tol, M.B, Hovius, R, Graff, A, Stahlberg, H, Tomizaki, T, Desmyter, A, Moreau, C, Li, X.-D, Poitevin, F, Vogel, H, Nury, H. | | Deposit date: | 2014-05-09 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | X-ray structure of the mouse serotonin 5-HT3 receptor.
Nature, 512, 2014
|
|
3QXL
 
 | | Crystal structure of the CDC25 Domain from Ral-specific Guanine-nucleotide Exchange Factor RalGPS1a | | Descriptor: | Ras-specific guanine nucleotide-releasing factor RalGPS1 | | Authors: | Peng, W, Xu, J, Guan, X, Sun, Y, Li, X, Zhang, X.C, Rao, Z. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.237 Å) | | Cite: | Structural study of the Cdc25 domain from Ral-specific guanine-nucleotide exchange factor RalGPS1a.
Protein Cell, 2, 2011
|
|
1YNQ
 
 | | aldo-keto reductase AKR11C1 from Bacillus halodurans (holo form) | | Descriptor: | GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, ... | | Authors: | Marquardt, T, Kostrewa, D, Winkler, F.K, Li, X.D. | | Deposit date: | 2005-01-25 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution Crystal Structure of AKR11C1 from Bacillus halodurans: An NADPH-dependent 4-Hydroxy-2,3-trans-nonenal Reductase
J.Mol.Biol., 354, 2005
|
|
1YNP
 
 | | aldo-keto reductase AKR11C1 from Bacillus halodurans (apo form) | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Marquardt, T, Kostrewa, D, Winkler, F.K, Li, X.D. | | Deposit date: | 2005-01-25 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High-resolution Crystal Structure of AKR11C1 from Bacillus halodurans: An NADPH-dependent 4-Hydroxy-2,3-trans-nonenal Reductase
J.Mol.Biol., 354, 2005
|
|
4FI1
 
 | | Crystal structure of scCK2 alpha in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, MAGNESIUM ION, ... | | Authors: | Liu, H, Wang, H, Teng, M, Li, X. | | Deposit date: | 2012-06-07 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of scCK2 alpha in complex with ATP
To be Published
|
|
1ZTY
 
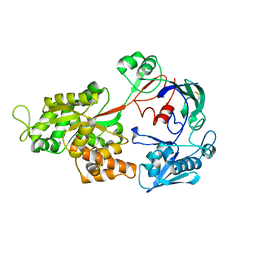 | |
1ZU0
 
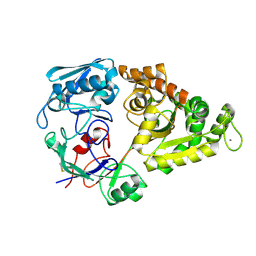 | |
6J60
 
 | | hnRNP A1 reversible amyloid core GFGGNDNFG (residues 209-217) | | Descriptor: | 9-mer peptide (GFGGNDNFG) from Heterogeneous nuclear ribonucleoprotein A1 | | Authors: | Luo, F, Zhou, H, Gui, X, Li, D, Li, X, Liu, C. | | Deposit date: | 2019-01-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.96 Å) | | Cite: | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
6IY3
 
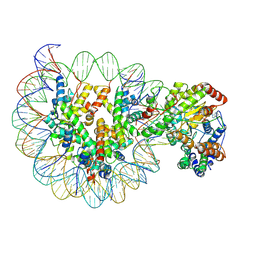 | | Structure of Snf2-MMTV-A nucleosome complex at shl-2 in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (147-MER), Histone H2A, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
6IY2
 
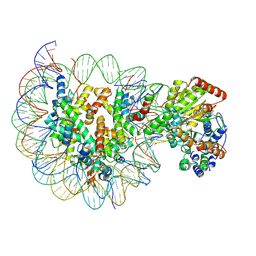 | | Structure of Snf2-MMTV-A nucleosome complex at shl2 in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (147-MER), DNA (167-MER), ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
4YLY
 
 | | Crystal structure of peptidyl-tRNA hydrolase from a Gram-positive bacterium, staphylococcus aureus at 2.25 angstrom resolution | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Zhang, F, Song, Y, Li, X, Teng, M.K. | | Deposit date: | 2015-03-06 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of Staphylococcus aureus peptidyl-tRNA hydrolase at a 2.25 angstrom resolution.
Acta Biochim.Biophys.Sin., 47, 2015
|
|
2GO4
 
 | | Crystal structure of Aquifex aeolicus LpxC complexed with TU-514 | | Descriptor: | 1,5-ANHYDRO-2-C-(CARBOXYMETHYL-N-HYDROXYAMIDE)-2-DEOXY-3-O-MYRISTOYL-D-GLUCITOL, CHLORIDE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Gennadios, H.A, Whittington, D.A, Li, X, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2006-04-12 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanistic Inferences from the Binding of Ligands to LpxC, a Metal-Dependent Deacetylase
Biochemistry, 45, 2006
|
|
5ERG
 
 | | Crystal structure of the two-subunit tRNA m1A58 methyltransferase TRM6-TRM61 in complex with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRM61, tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 | | Authors: | Zhu, Y, Wang, M, Wang, C, Fan, X, Jiang, X, Teng, M, Li, X. | | Deposit date: | 2015-11-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Crystal structure of the two-subunit tRNA m(1)A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae.
Sci Rep, 6, 2016
|
|
5EQJ
 
 | | Crystal structure of the two-subunit tRNA m1A58 methyltransferase from Saccharomyces cerevisiae | | Descriptor: | tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRM61, tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 | | Authors: | Zhu, Y, Wang, M, Wang, C, Fan, X, Jiang, X, Teng, M, Li, X. | | Deposit date: | 2015-11-13 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the two-subunit tRNA m(1)A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae.
Sci Rep, 6, 2016
|
|
7DCY
 
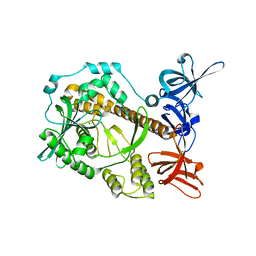 | | Apo form of Mycoplasma genitalium RNase R | | Descriptor: | MAGNESIUM ION, Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-10-27 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
7DIC
 
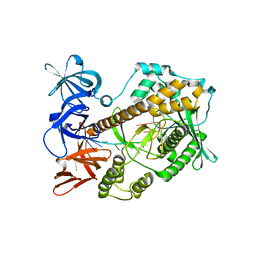 | | Mycoplasma genitalium RNase R in complex with single-stranded RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-11-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
7DOL
 
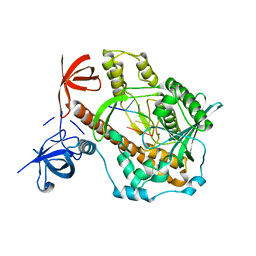 | | Mycoplasma genitalium RNase R in complex with double-stranded RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
7DID
 
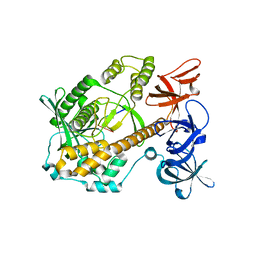 | | Mycoplasma genitalium RNase R in complex with ribose methylated single-stranded RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*AP*AP*AP*A)-3'), Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-11-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
7DNU
 
 | | mRNA-decapping enzyme g5Rp with inhibitor insp6 complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, mRNA-decapping protein g5R | | Authors: | Yang, Y, Chen, C, Li, L, Li, X.H, Su, D. | | Deposit date: | 2020-12-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Structural Insight into Molecular Inhibitory Mechanism of InsP 6 on African Swine Fever Virus mRNA-Decapping Enzyme g5Rp.
J.Virol., 96, 2022
|
|
7DNT
 
 | | mRNA-decapping enzyme g5Rp | | Descriptor: | mRNA-decapping protein g5R | | Authors: | Yang, Y, Chen, C, Li, L, Li, X.H, Su, D. | | Deposit date: | 2020-12-10 | | Release date: | 2022-03-09 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insight into Molecular Inhibitory Mechanism of InsP 6 on African Swine Fever Virus mRNA-Decapping Enzyme g5Rp.
J.Virol., 96, 2022
|
|
