2R1Z
 
 | |
5UGK
 
 | | Zinc-Binding Structure of a Catalytic Amyloid from Solid-State NMR Spectroscopy | | Descriptor: | ILE-HIS-VAL-HIS-LEU-GLN-ILE, ZINC ION | | Authors: | Lee, M, Wang, T, Makhlynets, O.V, Wu, Y, Polizzi, N, Wu, H, Gosavi, P.M, Korendovych, I.V, DeGrado, W.F, Hong, M. | | Deposit date: | 2017-01-09 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Zinc-binding structure of a catalytic amyloid from solid-state NMR.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1MFY
 
 | | SOLUTION STRUCTURE OF INFLUENZA A VIRUS C4 PROMOTER | | Descriptor: | C4 promoter of influneza A virus | | Authors: | Lee, M.-K, Bae, S.-H, Park, C.-J, Cheong, H.-K, Cheong, C, Choi, B.-S. | | Deposit date: | 2002-08-14 | | Release date: | 2002-09-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A single-nucleotide natural variation (U4 to C4) in an influenza A virus promoter exhibits a large structural change: implications for differential viral RNA synthesis by RNA-dependent RNA polymerase.
Nucleic Acids Res., 31, 2003
|
|
3DSQ
 
 | |
1XGE
 
 | | Dihydroorotase from Escherichia coli: Loop Movement and Cooperativity between subunits | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Chan, C.W, Guss, J.M, Christopherson, R.I, Maher, M.J. | | Deposit date: | 2004-09-17 | | Release date: | 2005-04-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dihydroorotase from Escherichia coli: Loop Movement and Cooperativity between Subunits
J.Mol.Biol., 348, 2005
|
|
5VDF
 
 | |
4MF9
 
 | |
4MGF
 
 | |
3HQ2
 
 | | BsuCP Crystal Structure | | Descriptor: | Bacillus subtilis M32 carboxypeptidase, CHLORIDE ION, FLUORIDE ION, ... | | Authors: | Lee, M.M, Isaza, C.E, White, J.D, Chen, R.P.-Y, Liang, G.F.-C, He, H.T.-F, Chan, S.I, Chan, M.K. | | Deposit date: | 2009-06-05 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insight into the substrate length restriction of M32 carboxypeptidases: Characterization of two distinct subfamilies.
Proteins, 77, 2009
|
|
2GW0
 
 | | A D(TGGGGT)- sodium and calcium complex. | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*T)-3', CALCIUM ION, SODIUM ION | | Authors: | Lee, M.P.H, Parkinson, G.N, Neidle, S. | | Deposit date: | 2006-05-03 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Observation of the coexistence of sodium and calcium ions in a DNA G-quadruplex ion channel.
J.Am.Chem.Soc., 129, 2007
|
|
3HOA
 
 | |
2EG7
 
 | | The crystal structure of E. coli dihydroorotase complexed with HDDP | | Descriptor: | 2-OXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4,6-DICARBOXYLIC ACID, Dihydroorotase, ZINC ION | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-02-28 | | Release date: | 2007-07-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Ligand-free and Inhibitor Complexes of Dihydroorotase from Escherichia coli: Implications for Loop Movement in Inhibitor Design
J.Mol.Biol., 370, 2007
|
|
2EG6
 
 | |
2EG8
 
 | | The crystal structure of E. coli dihydroorotase complexed with 5-fluoroorotic acid | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, ZINC ION | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-02-28 | | Release date: | 2007-07-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Ligand-free and Inhibitor Complexes of Dihydroorotase from Escherichia coli: Implications for Loop Movement in Inhibitor Design
J.Mol.Biol., 370, 2007
|
|
2LWK
 
 | | Solution structure of small molecule-influenza RNA complex | | Descriptor: | 6,7-dimethoxy-2-(piperazin-1-yl)quinazolin-4-amine, RNA (32-MER) | | Authors: | Lee, M.-K, Varani, G, Choi, B.-S, Pellecchia, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-08-01 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel small-molecule binds to the influenza A virus RNA promoter and inhibits viral replication.
Chem.Commun.(Camb.), 50, 2014
|
|
5VDE
 
 | |
3TL1
 
 | | Crystal structure of the Streptomyces coelicolor WhiE ORFVI polyketide aromatase/cyclase | | Descriptor: | 6,7,9-trihydroxy-3-methyl-1H-benzo[g]isochromen-1-one, GLYCEROL, Polyketide cyclase | | Authors: | Lee, M.-Y, Ames, B.D, Zhang, W, Tang, Y, Tsai, S.-C. | | Deposit date: | 2011-08-29 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insight into the Molecular Basis of Aromatic Polyketide Cyclization: Crystal Structure and in Vitro Characterization of WhiE-ORFVI.
Biochemistry, 51, 2012
|
|
2AAN
 
 | |
7F3V
 
 | | Crystal structure of YfiH with C107A mutation in complex with endogenous UDP-MurNAc | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, ... | | Authors: | Lee, M.S, Hsieh, K.Y, Chang, C.I. | | Deposit date: | 2021-06-17 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Basis for the Peptidoglycan-Editing Activity of YfiH.
Mbio, 13, 2021
|
|
7JGV
 
 | | CRYSTAL STRUCTURE OF BCL-XL IN COMPLEX WITH COMPOUND 1620116, CRYSTAL FORM 2 | | Descriptor: | 6-[(8E)-8-{2-[4-(benzylcarbamoyl)-1,3-thiazol-2-yl]hydrazinylidene}-5,6,7,8-tetrahydronaphthalen-2-yl]-3-(2-phenylethoxy)pyridine-2-carboxylic acid, Bcl-2-like protein 1 | | Authors: | Lee, M, Fairlie, W.D, Smith, B.J, Lee, E.F. | | Deposit date: | 2020-07-19 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Optimization of Benzothiazole and Thiazole Hydrazones as Inhibitors of Schistosome BCL-2.
Acs Infect Dis., 7, 2021
|
|
7JGW
 
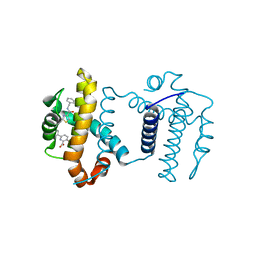 | | Crystal structure of BCL-XL in complex with COMPOUND 1620116, CRYSTAL FORM 1 | | Descriptor: | 6-[(8E)-8-{2-[4-(benzylcarbamoyl)-1,3-thiazol-2-yl]hydrazinylidene}-5,6,7,8-tetrahydronaphthalen-2-yl]-3-(2-phenylethoxy)pyridine-2-carboxylic acid, Bcl-2-like protein 1 | | Authors: | Lee, M, Fairlie, W.D, Smith, B.J, Lee, E.F. | | Deposit date: | 2020-07-19 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Optimization of Benzothiazole and Thiazole Hydrazones as Inhibitors of Schistosome BCL-2.
Acs Infect Dis., 7, 2021
|
|
2GYX
 
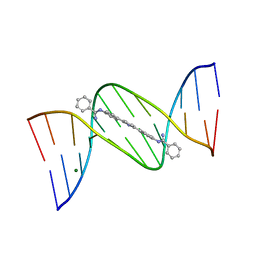 | | Crystal structure of DB884- D(CGCGAATTCGCG)2 complex at 1.86 A resolution. | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION, N,N'-(1H-PYRROLE-2,5-DIYLDI-4,1-PHENYLENE)DIBENZENECARBOXIMIDAMIDE | | Authors: | Lee, M.P.H, Neidle, S. | | Deposit date: | 2006-05-10 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Induced fit conformational changes of a "reversed amidine" heterocycle: optimized interactions in a DNA minor groove complex.
J.Am.Chem.Soc., 129, 2007
|
|
5WPA
 
 | | Structure of human SFPQ/PSPC1 heterodimer | | Descriptor: | Paraspeckle component 1, Splicing factor, proline- and glutamine-rich | | Authors: | Lee, M. | | Deposit date: | 2017-08-04 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of a SFPQ/PSPC1 heterodimer provides insights into preferential heterodimerization of human DBHS family proteins.
J. Biol. Chem., 293, 2018
|
|
2GWE
 
 | |
2GWQ
 
 | |
