7LHQ
 
 | | Solution structure of SARS-CoV-2 nonstructural protein 7 at pH 7.0 | | Descriptor: | Non-structural protein 7 | | Authors: | Lee, Y, Tonelli, M, Anderson, T.K, Kirchdoerfer, R.N, Henzler-Wildman, K, Lee, W. | | Deposit date: | 2021-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | pH-dependent polymorphism of the structure of SARS-CoV-2 nsp7
Biorxiv, 2021
|
|
8J0J
 
 | | AtSLAC1 8D mutant in closed state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8J1E
 
 | | AtSLAC1 in open state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2023-04-12 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
6LVT
 
 | |
6TKO
 
 | |
4XSJ
 
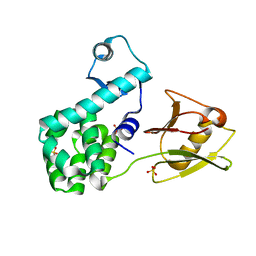 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
4XTB
 
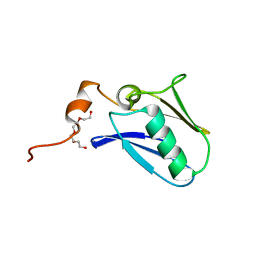 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter | | Descriptor: | Calcium uniporter protein, mitochondrial, TETRAETHYLENE GLYCOL | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
7QRU
 
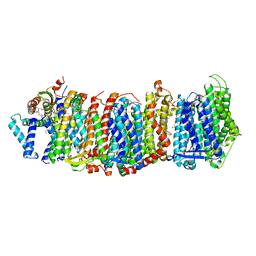 | | Structure of Bacillus pseudofirmus Mrp antiporter complex, monomer | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Na(+)/H(+) antiporter subunit B, Na(+)/H(+) antiporter subunit C, ... | | Authors: | Lee, Y. | | Deposit date: | 2022-01-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Ion transfer mechanisms in Mrp-type antiporters from high resolution cryoEM and molecular dynamics simulations.
Nat Commun, 13, 2022
|
|
8KHO
 
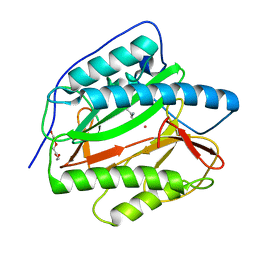 | | Crystal structure of human methionine aminopeptidase 12 (MAP12) in complex with two Cobalt ions and Methionine | | Descriptor: | COBALT (II) ION, METHIONINE, Methionine aminopeptidase 1D, ... | | Authors: | Lee, Y, Lee, E, Hahn, H, Kim, H, Heo, Y, Jang, D.M, Kim, H.J, Kim, H.S. | | Deposit date: | 2023-08-22 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into N-terminal methionine cleavage by the human mitochondrial methionine aminopeptidase, MetAP1D.
Sci Rep, 13, 2023
|
|
8KHN
 
 | | Crystal structure of human methionine aminopeptidase 12 (MAP12) in complex with two cobalt ions | | Descriptor: | COBALT (II) ION, Methionine aminopeptidase 1D, mitochondrial, ... | | Authors: | Lee, Y, Lee, E, Hahn, H, Kim, H, Heo, Y, Jang, D.M, Kim, H.J, Kim, H.S. | | Deposit date: | 2023-08-22 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural insights into N-terminal methionine cleavage by the human mitochondrial methionine aminopeptidase, MetAP1D.
Sci Rep, 13, 2023
|
|
8KHM
 
 | | Crystal structure of human methionine aminopeptidase 12 (MAP12) in the unbound form | | Descriptor: | GLYCEROL, Methionine aminopeptidase 1D, mitochondrial, ... | | Authors: | Lee, Y, Lee, E, Hahn, H, Kim, H, Heo, Y, Jang, D.M, Kim, H.J, Kim, H.S. | | Deposit date: | 2023-08-22 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural insights into N-terminal methionine cleavage by the human mitochondrial methionine aminopeptidase, MetAP1D.
Sci Rep, 13, 2023
|
|
8KA2
 
 | |
8K9Z
 
 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-bound calmodulin | | Descriptor: | CALCIUM ION, Calmodulin-2, RDTND-RID CBD | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
8KA1
 
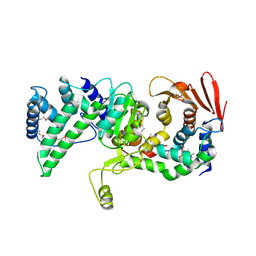 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-free calmodulin | | Descriptor: | Calmodulin-2, MAGNESIUM ION, RDTND-RID CBD | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
8KA0
 
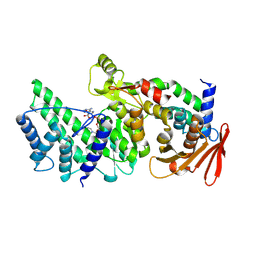 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-bound calmodulin and a nicotinamide adenine dinucleotide (NAD+) | | Descriptor: | CALCIUM ION, Calmodulin-2, GLYCEROL, ... | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
6LVU
 
 | |
5BZ6
 
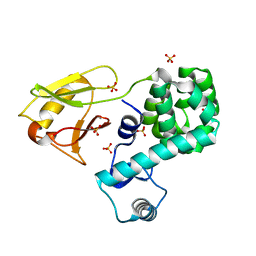 | | Crystal structure of the N-terminal domain single mutant (S92A) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-06-11 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
8GW7
 
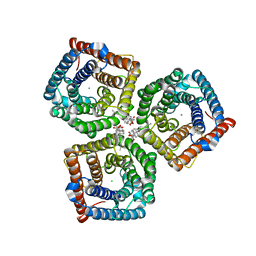 | | AtSLAC1 6D mutant in open state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein (Fragment) | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2022-09-16 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8GW6
 
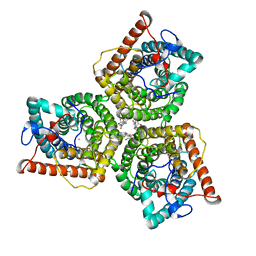 | | AtSLAC1 6D mutant in closed state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein (Fragment) | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2022-09-16 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
7NF8
 
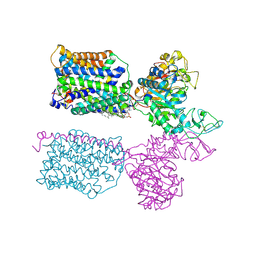 | | Ovine (b0,+AT-rBAT)2 hetero-tetramer, asymmetric unit, rigid-body fitted | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, Y, Kuehlbrandt, W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Ca 2+ -mediated higher-order assembly of heterodimers in amino acid transport system b 0,+ biogenesis and cystinuria.
Nat Commun, 13, 2022
|
|
7NF7
 
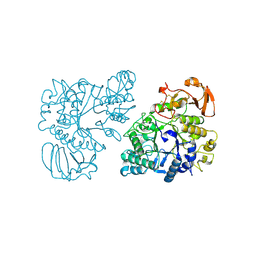 | | Ovine rBAT ectodomain homodimer, asymmetric unit | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lee, Y, Kuehlbrandt, W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-01-19 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Ca 2+ -mediated higher-order assembly of heterodimers in amino acid transport system b 0,+ biogenesis and cystinuria.
Nat Commun, 13, 2022
|
|
7NF6
 
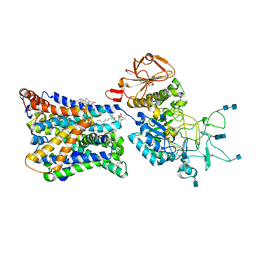 | | Ovine b0,+AT-rBAT heterodimer | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, Y, Kuehlbrandt, W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-01-19 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Ca 2+ -mediated higher-order assembly of heterodimers in amino acid transport system b 0,+ biogenesis and cystinuria.
Nat Commun, 13, 2022
|
|
5TSP
 
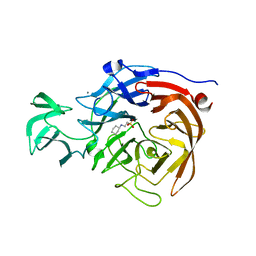 | | Crystal structure of the catalytic domain of Clostridium perfringens neuraminidase (NanI) in complex with a CHES | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, Sialidase | | Authors: | Lee, Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Kang, J.Y, Jin, M.S, Ryu, Y.B, Park, K.H, Eom, S.H. | | Deposit date: | 2016-10-31 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of the catalytic domain of Clostridium perfringens neuraminidase in complex with a non-carbohydrate-based inhibitor, 2-(cyclohexylamino)ethanesulfonic acid
Biochem. Biophys. Res. Commun., 486, 2017
|
|
4X5M
 
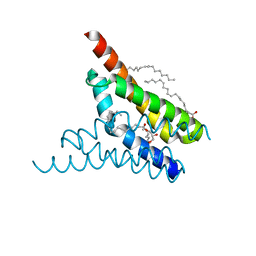 | | Crystal structure of SemiSWEET in the inward-open conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, OLEIC ACID, ... | | Authors: | Lee, Y, Nishizawa, T, Yamashita, K, Ishitani, R, Nureki, O. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the facilitative diffusion mechanism by SemiSWEET transporter
Nat Commun, 6, 2015
|
|
4X5N
 
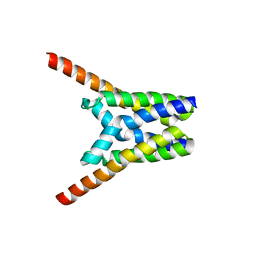 | | Crystal structure of SemiSWEET in the inward-open and outward-open conformations | | Descriptor: | Uncharacterized protein | | Authors: | Lee, Y, Nishizawa, T, Yamashita, K, Ishitani, R, Nureki, O. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the facilitative diffusion mechanism by SemiSWEET transporter
Nat Commun, 6, 2015
|
|
