3UXU
 
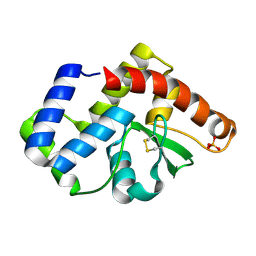 | |
5EQW
 
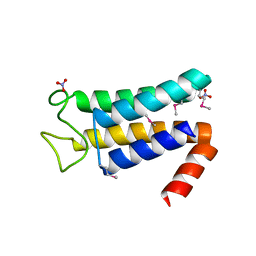 | | Structure of the major structural protein D135 of Acidianus tailed spindle virus (ATSV) | | Descriptor: | NITRATE ION, Putative major coat protein | | Authors: | Hochstein, R.A, Lintner, N.G, Young, M.J, Lawrence, C.M. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-16 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Structural studies ofAcidianustailed spindle virus reveal a structural paradigm used in the assembly of spindle-shaped viruses.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1R7I
 
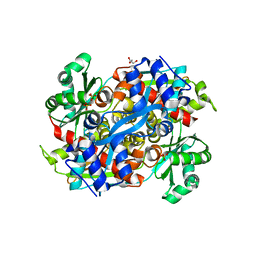 | | HMG-CoA Reductase from P. mevalonii, native structure at 2.2 angstroms resolution. | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase, GLYCEROL, SULFATE ION | | Authors: | Watson, J.M, Steussy, C.N, Burgner, J.W, Lawrence, C.M, Tabernero, L, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2003-10-21 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Investigations of the Basis for Stereoselectivity from the Binary Complex of HMG-COA Reductase.
To be Published
|
|
1R31
 
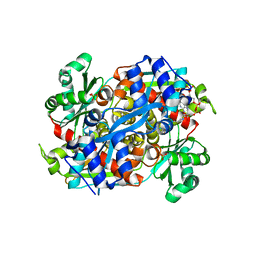 | | HMG-CoA reductase from Pseudomonas mevalonii complexed with HMG-CoA | | Descriptor: | (R)-MEVALONATE, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, COENZYME A, ... | | Authors: | Watson, J.M, Steussy, C.N, Burgner, J.W, Lawrence, C.M, Tabernero, L, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2003-09-30 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Investigations of the Basis for Stereoselectivity from the Binary Complex of HMG-CoA Reductase.
To be Published
|
|
1SKV
 
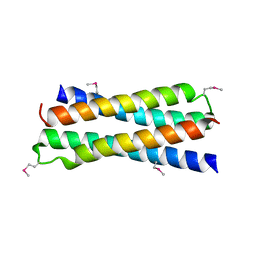 | | Crystal Structure of D-63 from Sulfolobus Spindle Virus 1 | | Descriptor: | Hypothetical 7.5 kDa protein | | Authors: | Kraft, P, Kummel, D, Oeckinghaus, A, Gauss, G.H, Wiedenheft, B, Young, M, Lawrence, C.M. | | Deposit date: | 2004-03-05 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of d-63 from sulfolobus spindle-shaped virus 1: surface properties of the dimeric four-helix bundle suggest an adaptor protein function
J.Virol., 78, 2004
|
|
1TBX
 
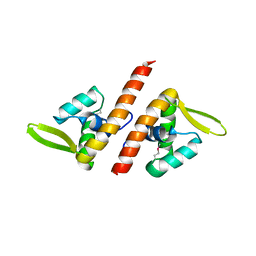 | | Crystal structure of SSV1 F-93 | | Descriptor: | Hypothetical 11.0 kDa protein | | Authors: | Kraft, P, Oeckinghaus, A, Kummel, D, Gauss, G.H, Wiedenheft, B, Young, M, Lawrence, C.M. | | Deposit date: | 2004-05-20 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of F-93 from Sulfolobus spindle-shaped virus 1, a winged-helix DNA binding protein.
J.Virol., 78, 2004
|
|
4AAI
 
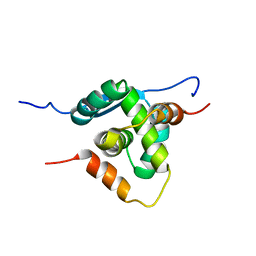 | | THERMOSTABLE PROTEIN FROM HYPERTHERMOPHILIC VIRUS SSV-RH | | Descriptor: | ORF E73 | | Authors: | Schlenker, C, Goel, A, Tripet, B.P, Menon, S, Lawrence, C.M, Copie, V. | | Deposit date: | 2011-12-02 | | Release date: | 2012-01-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Studies of E73 from a Hyperthermophilic Archaeal Virus Identify the "Rh3" Domain, an Elaborated Ribbon-Helix- Helix Motif Involved in DNA Recognition.
Biochemistry, 51, 2012
|
|
1U6A
 
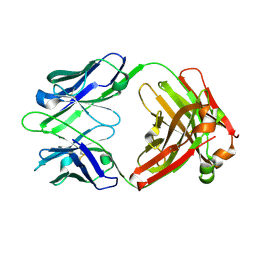 | | Crystal Structure of the Broadly Neutralizing Anti-HIV Fab F105 | | Descriptor: | F105 HEAVY CHAIN, F105 LIGHT CHAIN | | Authors: | Wilkinson, R.A, Piscitelli, C, Teintze, M, Lawrence, C.M. | | Deposit date: | 2004-07-29 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure of the Fab fragment of F105, a broadly reactive anti-human immunodeficiency virus (HIV) antibody that recognizes the CD4 binding site of HIV type 1 gp120.
J.Virol., 79, 2005
|
|
6VJG
 
 | | Csx3-I222 Crystal Form at 1.8 Angstrom Resolution | | Descriptor: | CRISPR-associated protein, Csx3 family | | Authors: | Brown, S, Charbonneau, A, Burman, N, Gauvin, C.C, Lawrence, C.M. | | Deposit date: | 2020-01-15 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Csx3 is a cyclic oligonucleotide phosphodiesterase associated with type III CRISPR-Cas that degrades the second messenger cA 4 .
J.Biol.Chem., 295, 2020
|
|
6W11
 
 | |
