7B5E
 
 | | Structure of calcium-bound mTMEM16A(ac)-I551A chloride channel at 4.1 A resolution | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Lam, A.K.M, Rheinberger, J, Paulino, C, Dutzler, R. | | Deposit date: | 2020-12-03 | | Release date: | 2021-02-10 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Gating the pore of the calcium-activated chloride channel TMEM16A.
Nat Commun, 12, 2021
|
|
7B5C
 
 | | Structure of calcium-bound mTMEM16A(ac) chloride channel at 3.7 A resolution | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Lam, A.K.M, Rheinberger, J, Paulino, C, Dutzler, R. | | Deposit date: | 2020-12-03 | | Release date: | 2021-02-10 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Gating the pore of the calcium-activated chloride channel TMEM16A.
Nat Commun, 12, 2021
|
|
7B5D
 
 | |
7ZK3
 
 | | Structure of 1PBC- and calcium-bound mTMEM16A(ac) chloride channel at 2.85 A resolution | | Descriptor: | 1-Hydroxy-3-(trifluoromethyl)pyrido[1,2-a]benzimidazole-4-carbonitrile, Anoctamin-1, CALCIUM ION | | Authors: | Lam, A.K.M, Rutz, S, Dutzler, R. | | Deposit date: | 2022-04-12 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Inhibition mechanism of the chloride channel TMEM16A by the pore blocker 1PBC.
Nat Commun, 13, 2022
|
|
3CFI
 
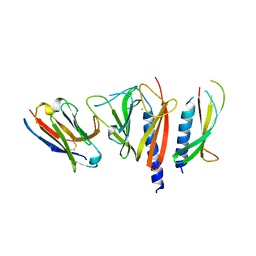 | | Nanobody-aided structure determination of the EPSI:EPSJ pseudopilin heterdimer from Vibrio Vulnificus | | Descriptor: | CHLORIDE ION, Nanobody NBEPSIJ_11, Type II secretory pathway, ... | | Authors: | Lam, A.Y, Pardon, E, Korotkov, K.V, Steyaert, J, Hol, W.G.J. | | Deposit date: | 2008-03-03 | | Release date: | 2009-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Nanobody-aided structure determination of the EpsI:EpsJ pseudopilin heterodimer from Vibrio vulnificus.
J.Struct.Biol., 166, 2009
|
|
8QZC
 
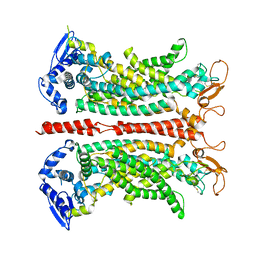 | |
1QDS
 
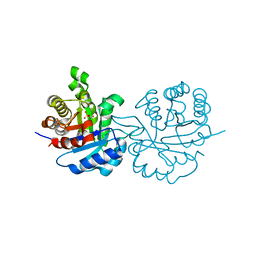 | | SUPERSTABLE E65Q MUTANT OF LEISHMANIA MEXICANA TRIOSEPHOSPHATE ISOMERASE (TIM) | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Lambeir, A.M, Backmann, J, Ruiz-Sanz, J, Filimonov, V, Nielsen, J.E, Vriend, G, Kursula, I, Norledge, B.V, Wierenga, R.K. | | Deposit date: | 1999-07-10 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The ionization of a buried glutamic acid is thermodynamically linked to the stability of Leishmania mexicana triose phosphate isomerase.
Eur.J.Biochem., 267, 2000
|
|
1QUP
 
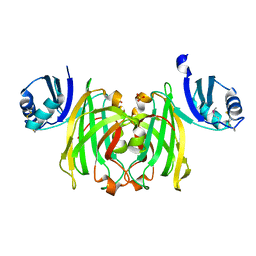 | | CRYSTAL STRUCTURE OF THE COPPER CHAPERONE FOR SUPEROXIDE DISMUTASE | | Descriptor: | SULFATE ION, SUPEROXIDE DISMUTASE 1 COPPER CHAPERONE | | Authors: | Lamb, A.L, Wernimont, A.K, Pufahl, R.A, O'Halloran, T.V, Rosenzweig, A.C. | | Deposit date: | 1999-07-01 | | Release date: | 1999-12-10 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the copper chaperone for superoxide dismutase.
Nat.Struct.Biol., 6, 1999
|
|
4TYY
 
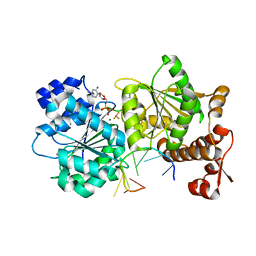 | | DEAD-box helicase Mss116 bound to ssRNA and CDP-BeF | | Descriptor: | ATP-dependent RNA helicase MSS116, mitochondrial, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Mallam, A.L, Sidote, D.J, Lambowitz, A.M. | | Deposit date: | 2014-07-09 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Elife, 3, 2014
|
|
6W3K
 
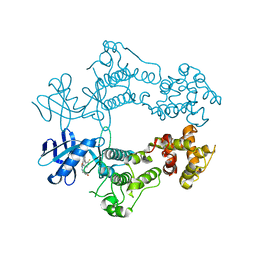 | | Structure of unphosphorylated human IRE1 bound to G-9807 | | Descriptor: | 4-[5-[2,6-bis(fluoranyl)phenyl]-2~{H}-pyrazolo[3,4-b]pyridin-3-yl]-2-(4-oxidanylpiperidin-1-yl)-1~{H}-pyrimidin-6-one, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Lammens, A, Wang, W, Ferri, E, Rudolph, J. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
4TZ0
 
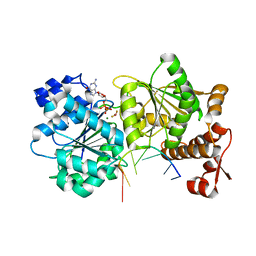 | | DEAD-box helicase Mss116 bound to ssRNA and GDP-BeF | | Descriptor: | ATP-dependent RNA helicase MSS116, mitochondrial, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Mallam, A.L, Sidote, D.J, Lambowitz, A.M. | | Deposit date: | 2014-07-09 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Elife, 3, 2014
|
|
4TYW
 
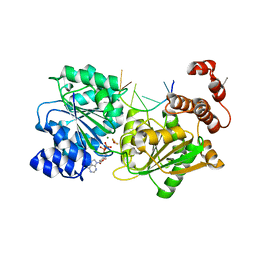 | | DEAD-box helicase Mss116 bound to ssRNA and ADP-BeF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase MSS116, mitochondrial, ... | | Authors: | Mallam, A.L, Sidote, D.J, Lambowitz, A.M. | | Deposit date: | 2014-07-09 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Elife, 3, 2014
|
|
4TZ6
 
 | | DEAD-box helicase Mss116 bound to ssRNA and UDP-BeF | | Descriptor: | ATP-dependent RNA helicase MSS116, mitochondrial, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Mallam, A.L, Sidote, D.J, Lambowitz, A.M. | | Deposit date: | 2014-07-09 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Elife, 3, 2014
|
|
4TYN
 
 | | DEAD-box helicase Mss116 bound to ssDNA and ADP-BeF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase MSS116, mitochondrial, ... | | Authors: | Mallam, A.L, Sidote, D.J, Lambowitz, A.M. | | Deposit date: | 2014-07-08 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.959 Å) | | Cite: | Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Elife, 3, 2014
|
|
4DB4
 
 | | Mss116p DEAD-box helicase domain 2 bound to a chimaeric RNA-DNA duplex | | Descriptor: | 5'-R(*GP*GP*GP*CP*GP*GP*G)-D(P*CP*CP*CP*GP*CP*CP*C)-3', ATP-dependent RNA helicase MSS116, mitochondrial | | Authors: | Mallam, A.L, Del Campo, M, Gilman, B.D, Sidote, D.J, Lambowitz, A. | | Deposit date: | 2012-01-13 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.599 Å) | | Cite: | Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p.
Nature, 490, 2012
|
|
4DB2
 
 | | Mss116p DEAD-box helicase domain 2 bound to an RNA duplex | | Descriptor: | 5'-R(*GP*GP*GP*CP*GP*GP*GP*CP*CP*CP*GP*CP*CP*C)-3', ATP-dependent RNA helicase MSS116, mitochondrial | | Authors: | Mallam, A.L, Del Campo, M, Gilman, B.D, Sidote, D.J, Lambowitz, A. | | Deposit date: | 2012-01-13 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.157 Å) | | Cite: | Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p.
Nature, 490, 2012
|
|
6GL3
 
 | | Crystal structure of human Phosphatidylinositol 4-kinase III beta (PI4KIIIbeta) in complex with ligand 44 | | Descriptor: | (3~{S})-4-(6-azanyl-1-methyl-pyrazolo[3,4-d]pyrimidin-4-yl)-~{N}-(4-methoxy-2-methyl-phenyl)-3-methyl-piperazine-1-carboxamide, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta | | Authors: | Lammens, A, Augustin, M, Steinbacher, S, Reuberson, J. | | Deposit date: | 2018-05-22 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Discovery of a Potent, Orally Bioavailable PI4KIII beta Inhibitor (UCB9608) Able To Significantly Prolong Allogeneic Organ Engraftment in Vivo.
J. Med. Chem., 61, 2018
|
|
3KTA
 
 | |
6FN4
 
 | | Apo form of UIC2 Fab complex of human-mouse chimeric ABCB1 (ABCB1HM) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Apo form of Human-mouse chimeric ABCB1 (ABCB1HM) in complex with Antigen binding fragment of UIC2., ... | | Authors: | Alam, A, Locher, K.P. | | Deposit date: | 2018-02-02 | | Release date: | 2018-02-21 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structure of a zosuquidar and UIC2-bound human-mouse chimeric ABCB1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FN1
 
 | | Zosuquidar and UIC2 Fab complex of human-mouse chimeric ABCB1 (ABCB1HM) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Human-mouse chimeric ABCB1 (ABCBHM), UIC2 Antigen Binding Fragment Heavy Chain, ... | | Authors: | Alam, A, Locher, K.P. | | Deposit date: | 2018-02-02 | | Release date: | 2018-02-21 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structure of a zosuquidar and UIC2-bound human-mouse chimeric ABCB1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FXN
 
 | | Crystal structure of human BAFF in complex with Fab fragment of anti-BAFF antibody belimumab | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, belimumab heavy chain, belimumab light chain | | Authors: | Lammens, A, Maskos, K, Willen, L, Jiang, X, Schneider, P. | | Deposit date: | 2018-03-09 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A loop region of BAFF controls B cell survival and regulates recognition by different inhibitors.
Nat Commun, 9, 2018
|
|
7B1O
 
 | | Crystal structure of the indoleamine 2,3-dioxygenase 1 (IDO1) in complex with compound 22 | | Descriptor: | 4-chloranyl-N-[(1R)-1-[(1S,5R)-3-quinolin-4-yloxy-6-bicyclo[3.1.0]hexanyl]propyl]benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lammens, A, Krapp, S, Lewis, R.T, Hamilton, M.M. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme.
J.Med.Chem., 64, 2021
|
|
4ZCH
 
 | | Single-chain human APRIL-BAFF-BAFF Heterotrimer | | Descriptor: | TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, Tumor necrosis factor ligand superfamily member 13,Tumor necrosis factor ligand superfamily member 13B,Tumor necrosis factor ligand superfamily member 13B | | Authors: | Lammens, A, Jiang, X, Maskos, K, Schneider, P. | | Deposit date: | 2015-04-16 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Stoichiometry of Heteromeric BAFF and APRIL Cytokines Dictates Their Receptor Binding and Signaling Properties.
J.Biol.Chem., 290, 2015
|
|
4ZRP
 
 | | TC:CD320 | | Descriptor: | CALCIUM ION, CD320 antigen, CYANOCOBALAMIN, ... | | Authors: | Alam, A, Locher, K.P. | | Deposit date: | 2015-05-12 | | Release date: | 2016-07-20 | | Last modified: | 2021-08-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of transcobalamin recognition by human CD320 receptor.
Nat Commun, 7, 2016
|
|
4ZRQ
 
 | |
