3DLA
 
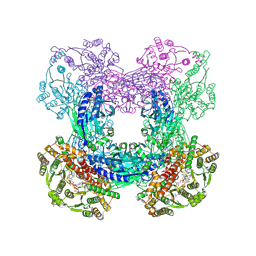 | | X-ray crystal structure of glutamine-dependent NAD+ synthetase from Mycobacterium tuberculosis bound to NaAD+ and DON | | Descriptor: | 5-OXO-L-NORLEUCINE, GLYCEROL, Glutamine-dependent NAD(+) synthetase, ... | | Authors: | LaRonde-LeBlanc, N.A, Resto, M, Gerratana, B. | | Deposit date: | 2008-06-26 | | Release date: | 2009-03-10 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Regulation of active site coupling in glutamine-dependent NAD(+) synthetase.
Nat.Struct.Mol.Biol., 16, 2009
|
|
1PUF
 
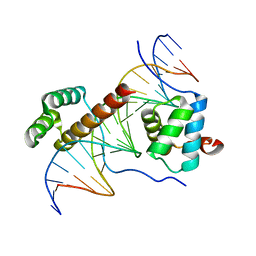 | | Crystal Structure of HoxA9 and Pbx1 homeodomains bound to DNA | | Descriptor: | 5'-D(*AP*CP*TP*CP*TP*AP*TP*GP*AP*TP*TP*TP*AP*CP*GP*AP*CP*GP*CP*T)-3', 5'-D(*TP*AP*GP*CP*GP*TP*CP*GP*TP*AP*AP*AP*TP*CP*AP*TP*AP*GP*AP*G)-3', Homeobox protein Hox-A9, ... | | Authors: | Laronde-Leblanc, N.A, Wolberger, C. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURE OF HOXA9 AND PBX1 BOUND TO DNA: HOX HEXAPEPTIDE AND DNA RECOGNITION ANTERIOR TO POSTERIOR
Genes Dev., 17, 2003
|
|
2NSZ
 
 | |
1TQM
 
 | |
1TQI
 
 | |
1TQP
 
 | |
1ZAO
 
 | | Crystal Structure of A.fulgidus Rio2 Kinase Complexed With ATP and Manganese Ions | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Laronde-Leblanc, N, Guszczynski, T, Copeland, T, Wlodawer, A. | | Deposit date: | 2005-04-06 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Autophosphorylation of Archaeoglobus fulgidus Rio2 and crystal structures of its nucleotide-metal ion complexes.
Febs J., 272, 2005
|
|
1ZAR
 
 | | Crystal Structure of A.fulgidus Rio2 Kinase Complexed With ADP and Manganese Ions | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Laronde-Leblanc, N, Guszczynski, T, Copeland, T, Wlodawer, A. | | Deposit date: | 2005-04-06 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Autophosphorylation of Archaeoglobus fulgidus Rio2 and crystal structures of its nucleotide-metal ion complexes.
Febs J., 272, 2005
|
|
3RE4
 
 | |
2IOL
 
 | |
2ION
 
 | |
2IOS
 
 | |
3O7B
 
 | |
3OII
 
 | |
3OIN
 
 | |
3OIJ
 
 | |
4JIN
 
 | | X-ray crystal structure of Archaeoglobus fulgidus Rio1 bound to (2E)-N-benzyl-2-cyano-3-(pyridine-4-yl)acrylamide (WP1086) | | Descriptor: | (2E)-N-benzyl-2-cyano-3-(pyridin-4-yl)prop-2-enamide, RIO-type serine/threonine-protein kinase Rio1 | | Authors: | Mielecki, M, Krawiec, K, Kiburu, I, Grzelak, K, Wlodzimierz, Z, Kierdaszuk, B, Kowa, K, Fokt, I, Szymanski, S, Piotr, S, Szeja, W, Priebe, W, Lesyng, B, LaRonde-LeBlanc, N. | | Deposit date: | 2013-03-06 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Development of novel molecular probes of the Rio1 atypical protein kinase.
Biochim.Biophys.Acta, 1834, 2013
|
|
4GYG
 
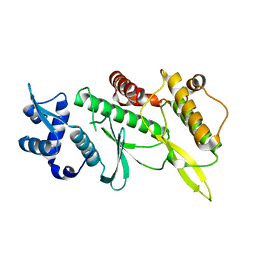 | | Crystal structure of the Rio2 kinase from Chaetomium thermophilum | | Descriptor: | Rio2 kinase | | Authors: | Ferreira-Cerca, S, Sagar, V, Schafer, T, Diop, M, Wesseling, A.M, Lu, H, Chai, E, Hurt, E, LaRonde-LeBlanc, N. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.482 Å) | | Cite: | ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4GYI
 
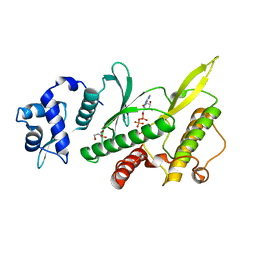 | | Crystal structure of the Rio2 kinase-ADP/Mg2+-phosphoaspartate complex from Chaetomium thermophilum | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ferreira-Cerca, S, Sagar, V, Schafer, T, Diop, M, Wesseling, A.M, Lu, H, Chai, E, Hurt, E, LaRonde-LeBlanc, N. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4KI8
 
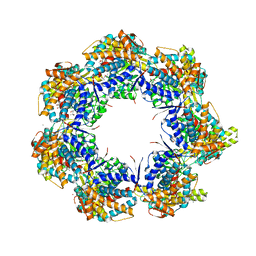 | | Crystal structure of a GroEL-ADP complex in the relaxed allosteric state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Fei, X, Yang, D, LaRonde-LeBlanc, N, Lorimer, G.H. | | Deposit date: | 2013-05-01 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.722 Å) | | Cite: | Crystal structure of a GroEL-ADP complex in the relaxed allosteric state at 2.7 A resolution.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3FRQ
 
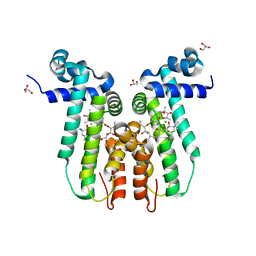 | | Structure of the macrolide biosensor protein, MphR(A), with erythromcyin | | Descriptor: | CHLORIDE ION, ERYTHROMYCIN A, GLYCEROL, ... | | Authors: | Zheng, J, Sagar, V, Smolinsky, A, Bourke, C, LaRonde-LeBlanc, N, Cropp, T.A. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure and function of the macrolide biosensor protein, MphR(A), with and without erythromycin
J.Mol.Biol., 387, 2009
|
|
3GH8
 
 | | Crystal structure of Mus musculus iodotyrosine deiodinase (IYD) bound to FMN and di-iodotyrosine (DIT) | | Descriptor: | 3,5-DIIODOTYROSINE, FLAVIN MONONUCLEOTIDE, Iodotyrosine dehalogenase 1, ... | | Authors: | Thomas, S.R, McTamney, P.M, Adler, J.M, LaRonde-LeBlanc, N, Rokita, S.E. | | Deposit date: | 2009-03-03 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of iodotyrosine deiodinase, a novel flavoprotein responsible for iodide salvage in thyroid glands.
J.Biol.Chem., 284, 2009
|
|
3GB5
 
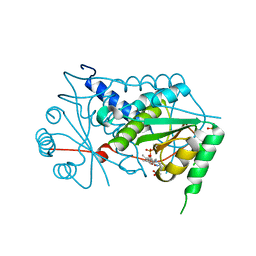 | | Crystal structure of Mus musculus iodotyrosine deiodinase (IYD) bound to FMN | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Iodotyrosine dehalogenase 1, ... | | Authors: | Thomas, S.R, McTamney, P.M, Adler, J.M, LaRonde-LeBlanc, N, Rokita, S.E. | | Deposit date: | 2009-02-18 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of iodotyrosine deiodinase, a novel flavoprotein responsible for iodide salvage in thyroid glands.
J.Biol.Chem., 284, 2009
|
|
3GFD
 
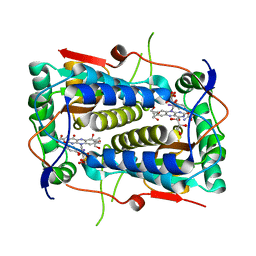 | | Crystal structure of Mus musculus iodotyrosine deiodinase (IYD) bound to FMN and mono-iodotyrosine (MIT) | | Descriptor: | 3-IODO-TYROSINE, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Thomas, S.R, McTamney, P.M, Adler, J.M, LaRonde-LeBlanc, N, Rokita, S.E. | | Deposit date: | 2009-02-26 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of iodotyrosine deiodinase, a novel flavoprotein responsible for iodide salvage in thyroid glands.
J.Biol.Chem., 284, 2009
|
|
3G56
 
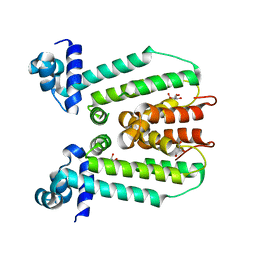 | | Structure of the macrolide biosensor protein, MphR(A) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Regulator of macrolide 2'-phosphotransferase I | | Authors: | Zheng, J, Sagar, V, Smolinsky, A, Bourke, C, LaRonde-LeBlanc, N, Cropp, T.A. | | Deposit date: | 2009-02-04 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of the macrolide biosensor protein, MphR(A), with and without erythromycin
J.Mol.Biol., 387, 2009
|
|
