6YM9
 
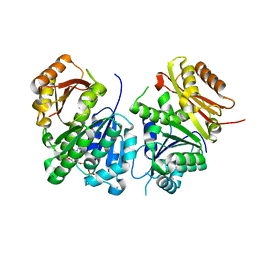 | | Mycobacterium tuberculosis FtsZ in complex with GTP-gamma-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell division protein FtsZ | | Authors: | Alnami, A.T, Norton, R.S, Pena, H.P, Haider, M, kozielski, F. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Conformational Flexibility of A Highly Conserved Helix Controls Cryptic Pocket Formation in FtsZ.
J.Mol.Biol., 433, 2021
|
|
2X7E
 
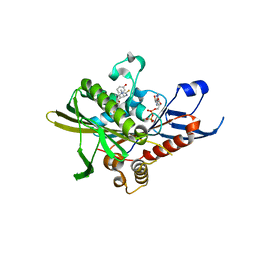 | | Crystal structure of human kinesin Eg5 in complex with (R)-fluorastrol | | Descriptor: | (4R)-5-[(S)-(3,4-DIFLUOROPHENYL)(HYDROXY)METHYL]-4-(3-HYDROXYPHENYL)-1,6-DIMETHYL-3,4-DIHYDROPYRIMIDINE-2(1H)-THIONE, ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KIF11, ... | | Authors: | Kaan, H.Y.K, Ulaganathan, V, Rath, O, Laggner, C, Prokopcova, H, Dallinger, D, Kappe, C.O, Kozielski, F. | | Deposit date: | 2010-02-26 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Inhibition of Eg5 by Dihydropyrimidines: Stereoselectivity of Antimitotic Inhibitors Enastron, Dimethylenastron and Fluorastrol.
J.Med.Chem., 53, 2010
|
|
8A55
 
 | |
8AZ8
 
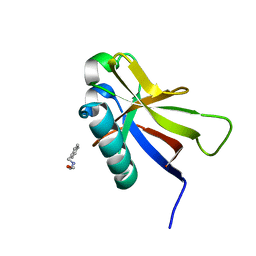 | | SARS-CoV-2 non-structural protein-1 (nsp1) in complex with 2-(benzylamino)ethan-1-ol | | Descriptor: | 2-[(phenylmethyl)amino]ethanol, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2022-09-05 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Two Ligand-Binding Sites on SARS-CoV-2 Non-Structural Protein 1 Revealed by Fragment-Based X-ray Screening.
Int J Mol Sci, 23, 2022
|
|
6H80
 
 | | Dengue-RdRp3-inhibitor complex co-crystallisation | | Descriptor: | 2-(4-methoxy-3-thiophen-2-yl-phenyl)ethanoic acid, DI(HYDROXYETHYL)ETHER, Genome polyprotein, ... | | Authors: | Talapatra, S.K, Kozielski, F. | | Deposit date: | 2018-07-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development and validation of RdRp Screen, a crystallization screen for viral RNA-dependent RNA polymerases.
Biol Open, 8, 2019
|
|
6H9R
 
 | | Dengue-RdRp3-inhibitor complex soaking | | Descriptor: | 2-(4-methoxy-3-thiophen-2-yl-phenyl)ethanoic acid, DI(HYDROXYETHYL)ETHER, Genome polyprotein, ... | | Authors: | Talapatra, S.K, Kozielski, F. | | Deposit date: | 2018-08-05 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development and validation of RdRp Screen, a crystallization screen for viral RNA-dependent RNA polymerases.
Biol Open, 8, 2019
|
|
6HKX
 
 | | Eg5-inhibitor complex | | Descriptor: | (2~{S})-2-(3-azanylpropyl)-5-[2,5-bis(fluoranyl)phenyl]-~{N}-methoxy-~{N}-methyl-2-phenyl-1,3,4-thiadiazole-3-carboxamide, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Talapatra, S.K, Kozielski, F. | | Deposit date: | 2018-09-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Is the Fate of Clinical Candidate Arry-520 Already Sealed? Predicting Resistance in Eg5-Inhibitor Complexes.
Mol.Cancer Ther., 18, 2019
|
|
6HKY
 
 | | Eg5-inhibitor complex | | Descriptor: | (2~{S})-2-(3-azanylpropyl)-5-[2,5-bis(fluoranyl)phenyl]-~{N}-methoxy-~{N}-methyl-2-phenyl-1,3,4-thiadiazole-3-carboxamide, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Talapatra, S.K, Kozielski, F. | | Deposit date: | 2018-09-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Is the Fate of Clinical Candidate Arry-520 Already Sealed? Predicting Resistance in Eg5-Inhibitor Complexes.
Mol.Cancer Ther., 18, 2019
|
|
6G6Y
 
 | | Eg5-inhibitor complex | | Descriptor: | (~{N}~{Z})-~{N}-[(5~{S})-4-ethanoyl-5-methyl-5-phenyl-1,3,4-thiadiazolidin-2-ylidene]ethanamide, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Talapatra, S.K, Kozielski, F, Tham, C.L. | | Deposit date: | 2018-04-03 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Eg5 - K858 complex and implications for structure-based design of thiadiazole-containing inhibitors.
Eur J Med Chem, 156, 2018
|
|
6HWS
 
 | | Keap1 - inhibitor complex | | Descriptor: | 1,2-ETHANEDIOL, 2-[[4-[2-hydroxy-2-oxoethyl-(4-methoxyphenyl)sulfonyl-amino]-3-phenylmethoxy-phenyl]-(4-methoxyphenyl)sulfonyl-amino]ethanoic acid, Kelch-like ECH-associated protein 1, ... | | Authors: | Talapatra, S.K, Kozielski, F, Wells, G, Georgakopoulos, N.D. | | Deposit date: | 2018-10-13 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Keap1-inhibitor complex
To Be Published
|
|
6ZCT
 
 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | Descriptor: | ZINC ION, nsp10 | | Authors: | Rogstam, A, Nyblom, M, Christensen, S, Sele, C, Lindvall, T, Rasmussen, A.A, Andre, I, Fisher, S.Z, Knecht, W, Kozielski, F. | | Deposit date: | 2020-06-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
7ORR
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00022 | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORV
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00239 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORU
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00221 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORW
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00265 | | Descriptor: | 1H-benzimidazol-4-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
2FE4
 
 | | The crystal structure of human neuronal Rab6B in its inactive GDP-bound form | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Garcia-Saez, I, Tcherniuk, F, Kozielski, F. | | Deposit date: | 2005-12-15 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of human neuronal Rab6B in the active and inactive form.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2X2R
 
 | | Crystal structure of human kinesin Eg5 in complex with (R)-2-amino-3-((4-chlorophenyl)diphenylmethylthio)propanoic acid | | Descriptor: | (2R)-2-AMINO-3-[(2R)-2-METHYL-1,1-DIPHENYL-BUTYL]SULFANYL-PROPANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KIF11, ... | | Authors: | Kaan, H.Y.K, Weiss, J, Menger, D, Ulaganathan, V, Laggner, C, Popowycz, F, Joseph, B, Kozielski, F. | | Deposit date: | 2010-01-15 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Activity Relationship and Multidrug Resistance Study of New S-Trityl-L-Cysteine Derivatives as Inhibitors of Eg5.
J.Med.Chem., 54, 2011
|
|
2XAE
 
 | | Crystal structure of human kinesin Eg5 in complex with (R)-2-amino-3-((S)-2-methyl-1,1-diphenylbutylthio)propanoic acid | | Descriptor: | (2R)-2-AMINO-3-[(4-CHLOROPHENYL)-DIPHENYL-METHYL]SULFANYL-PROPANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Kaan, H.Y.K, Weiss, J, Menger, D, Ulaganathan, V, Tkocz, K, Laggner, C, Popowycz, F, Joseph, B, Kozielski, F. | | Deposit date: | 2010-03-31 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Activity Relationship and Multidrug Resistance Study of New S-Trityl-L-Cysteine Derivatives as Inhibitors of Eg5.
J.Med.Chem., 54, 2011
|
|
2XT3
 
 | | HUMAN KIF7, A KINESIN INVOLVED IN HEDGEHOG SIGNALLING | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KIF7, MAGNESIUM ION | | Authors: | Klejnot, M, Kozielski, F. | | Deposit date: | 2010-10-04 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Structural Insights Into Human Kif7, a Kinesin Involved in Hedgehog Signalling.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2FFQ
 
 | | The crystal structure of human neuronal Rab6B in its active GTPgS-bound form | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-6B | | Authors: | Garcia-Saez, I, Tcherniuk, S, Kozielski, F. | | Deposit date: | 2005-12-20 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of human neuronal Rab6B in the active and inactive form.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4A51
 
 | |
2X7C
 
 | | Crystal structure of human kinesin Eg5 in complex with (S)-enastron | | Descriptor: | (S)-4-(3-HYDROXYPHENYL)-2-THIOXO-1,2,3,4,7,8-HEXAHYDROQUINAZOLIN-5(6H)-ONE, ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KIF11, ... | | Authors: | Kaan, H.Y.K, Ulaganathan, V, Rath, O, Laggner, C, Prokopcova, H, Dallinger, D, Kappe, C.O, Kozielski, F. | | Deposit date: | 2010-02-26 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Inhibition of Eg5 by Dihydropyrimidines: Stereoselectivity of Antimitotic Inhibitors Enastron, Dimethylenastron and Fluorastrol.
J.Med.Chem., 53, 2010
|
|
2X7D
 
 | | Crystal structure of human kinesin Eg5 in complex with (S)-dimethylenastron | | Descriptor: | (4S)-4-(3-HYDROXYPHENYL)-7,7-DIMETHYL-2-THIOXO-2,3,4,6,7,8-HEXAHYDROQUINAZOLIN-5(1H)-ONE, ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KIF11, ... | | Authors: | Kaan, H.Y.K, Ulaganathan, V, Rath, O, Laggner, C, Prokopcova, H, Dallinger, D, Kappe, C.O, Kozielski, F. | | Deposit date: | 2010-02-26 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Inhibition of Eg5 by Dihydropyrimidines: Stereoselectivity of Antimitotic Inhibitors Enastron, Dimethylenastron and Fluorastrol.
J.Med.Chem., 53, 2010
|
|
2WOG
 
 | | Intermediate and final states of human kinesin Eg5 in complex with S-trityl-L-cysteine | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KIF11, MAGNESIUM ION, ... | | Authors: | Kaan, H.Y.K, Ulaganathan, V, Hackney, D.D, Kozielski, F. | | Deposit date: | 2009-07-23 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Allosteric Transition Trapped in an Intermediate State of a New Kinesin-Inhibitor Complex.
Biochem.J., 425, 2010
|
|
2Y5W
 
 | |
