3HRR
 
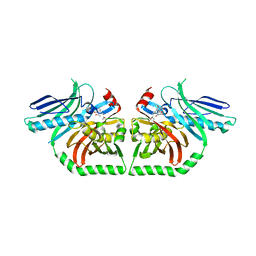 | | The Product Template Domain from PksA with Harris Compound Bound | | Descriptor: | 1-(3-acetyl-4,5-dihydroxy-7-methoxynaphthalen-2-yl)propan-2-one, Aflatoxin biosynthesis polyketide synthase | | Authors: | Korman, T.P, Tsai, S.C. | | Deposit date: | 2009-06-09 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for biosynthetic programming of fungal aromatic polyketide cyclization.
Nature, 461, 2009
|
|
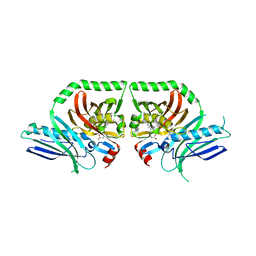
3HRQ
 
 | |
3ILS
 
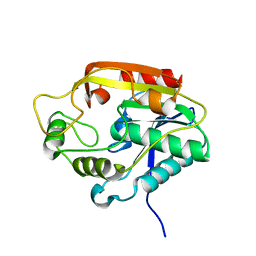 | | The Thioesterase Domain from PksA | | Descriptor: | Aflatoxin biosynthesis polyketide synthase | | Authors: | Korman, T.P. | | Deposit date: | 2009-08-07 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of an iterative polyketide synthase thioesterase domain catalyzing Claisen cyclization in aflatoxin biosynthesis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
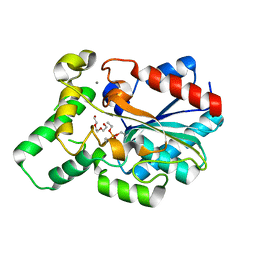
4GXN
 
 | |
4GW3
 
 | | Crystal Structure of the Lipase from Proteus mirabilis | | Descriptor: | CALCIUM ION, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Korman, T.P. | | Deposit date: | 2012-08-31 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Proteus mirabilis Lipase, a Novel Lipase from the Proteus/Psychrophilic Subfamily of Lipase Family I.1.
Plos One, 7, 2012
|
|
4HS9
 
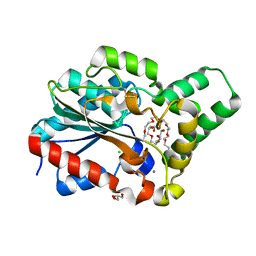 | | Methanol tolerant mutant of the Proteus mirabilis lipase | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Korman, T.P. | | Deposit date: | 2012-10-29 | | Release date: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dieselzymes: development of a stable and methanol tolerant lipase for biodiesel production by directed evolution.
Biotechnol Biofuels, 6, 2013
|
|
1X7G
 
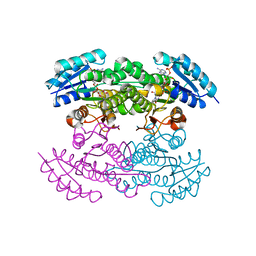 | | Actinorhodin Polyketide Ketoreductase, act KR, with NADP bound | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative ketoacyl reductase | | Authors: | Korman, T.P, Hill, J.A, Vu, T.N. | | Deposit date: | 2004-08-13 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of actinorhodin polyketide ketoreductase: cofactor binding and substrate specificity
Biochemistry, 43, 2004
|
|
1X7H
 
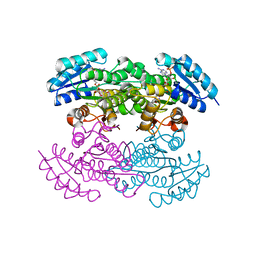 | | Actinorhodin Polyketide Ketoreductase, with NADPH bound | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative ketoacyl reductase | | Authors: | Korman, T.P, Hill, J.A, Vu, T.N. | | Deposit date: | 2004-08-13 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of actinorhodin polyketide ketoreductase: cofactor binding and substrate specificity
Biochemistry, 43, 2004
|
|
1XR3
 
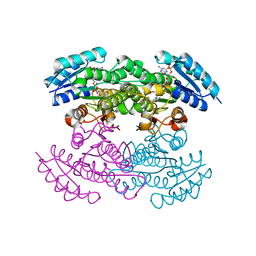 | | Actinorhodin Polyketide Ketoreductase with NADP and the Inhibitor Isoniazid bound | | Descriptor: | 4-(DIAZENYLCARBONYL)PYRIDINE, ACTINORHODIN POLYKETIDE KETOREDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Korman, T.P, Hill, J.A, Vu, T.N, Tsai, S.C. | | Deposit date: | 2004-10-13 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural analysis of actinorhodin polyketide ketoreductase: cofactor binding and substrate specificity.
Biochemistry, 43, 2004
|
|
3QRW
 
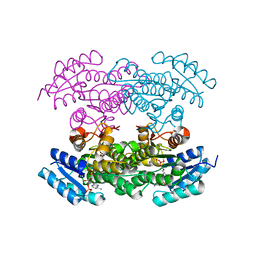 | |
2RH4
 
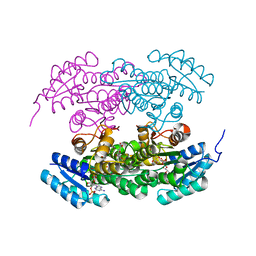 | | Actinorhodin ketoreductase, actKR, with NADPH and Inhibitor Emodin | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, Actinorhodin Polyketide Ketoreductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Korman, T.P, Tsai, S.-C. | | Deposit date: | 2007-10-05 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibition kinetics and emodin cocrystal structure of a type II polyketide ketoreductase
Biochemistry, 47, 2008
|
|
2RHC
 
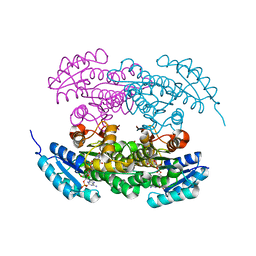 | | Actinorhodin ketordeuctase, actKR, with NADP+ and Inhibitor Emodin | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, Actinorhodin Polyketide Ketoreductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Korman, T.P, Tsai, S.-C. | | Deposit date: | 2007-10-08 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition kinetics and emodin cocrystal structure of a type II polyketide ketoreductase
Biochemistry, 47, 2008
|
|
2RHR
 
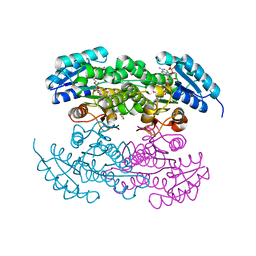 | | P94L actinorhodin ketordeuctase mutant, with NADPH and Inhibitor Emodin | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, Actinorhodin Polyketide Ketoreductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Korman, T.P, Tsai, S.-C. | | Deposit date: | 2007-10-09 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition kinetics and emodin cocrystal structure of a type II polyketide ketoreductase
Biochemistry, 47, 2008
|
|
6VGS
 
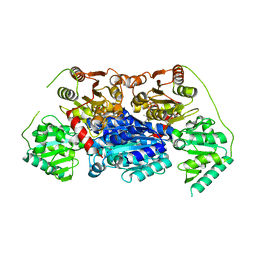 | | Alpha-ketoisovalerate decarboxylase (KivD) from Lactococcus lactis, thermostable mutant | | Descriptor: | Alpha-keto acid decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Chan, S, Korman, T.P, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2020-01-08 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isobutanol production freed from biological limits using synthetic biochemistry.
Nat Commun, 11, 2020
|
|
3NI3
 
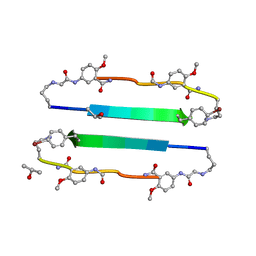 | | 54-Membered ring macrocyclic beta-sheet peptide | | Descriptor: | 54-membered ring macrocyclic beta-sheet peptide, ISOPROPYL ALCOHOL | | Authors: | Sawaya, M.R, Eisenberg, D, Nowick, J.S, Korman, T.P, Khakshoor, O. | | Deposit date: | 2010-06-15 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | X-ray crystallographic structure of an artificial beta-sheet dimer.
J.Am.Chem.Soc., 132, 2010
|
|
2RER
 
 | |
