1H8N
 
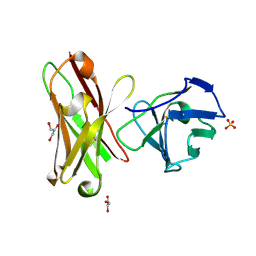 | | Three-dimensional structure of anti-ampicillin single chain Fv fragment from phage-displayed murine antibody libraries | | Descriptor: | GLYCEROL, MUTANT AL2 6E7S9G, SULFATE ION | | Authors: | Jung, S, Spinelli, S, Schimmele, B, Honegger, A, Pugliese, L, Cambillau, C, Pluckthun, A. | | Deposit date: | 2001-02-14 | | Release date: | 2001-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The Importance of Framework Residues H6, H7 and H10 in Antibody Heavy Chains: Experimental Evidence for a New Structural Subclassification of Antibody V(H) Domains
J.Mol.Biol., 309, 2001
|
|
1I3G
 
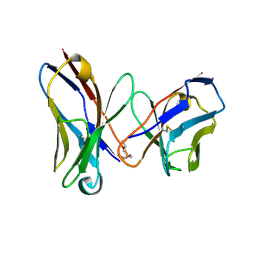 | | CRYSTAL STRUCTURE OF AN AMPICILLIN SINGLE CHAIN FV, FORM 1, FREE | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ANTIBODY FV FRAGMENT | | Authors: | Jung, S, Spinelli, S, Schimmele, B, Honegger, A, Pugliese, L, Cambillau, C, Pluckthun, A. | | Deposit date: | 2001-02-15 | | Release date: | 2001-10-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Selection, characterization and x-ray structure of anti-ampicillin single-chain Fv fragments from phage-displayed murine antibody libraries.
J.Mol.Biol., 309, 2001
|
|
3HB1
 
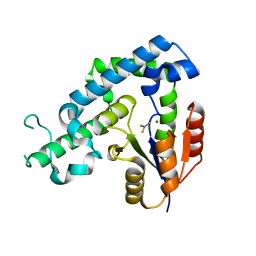 | | Crystal structure of ed-eya2 complexed with Alf3 | | Descriptor: | ALUMINUM FLUORIDE, Eyes absent homolog 2 (Drosophila), MAGNESIUM ION | | Authors: | Jung, S.K, Jeong, D.G, Ryu, S.E, Kim, S.J. | | Deposit date: | 2009-05-03 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of ED-Eya2: insight into dual roles as a protein tyrosine phosphatase and a transcription factor
Faseb J., 24, 2010
|
|
3HB0
 
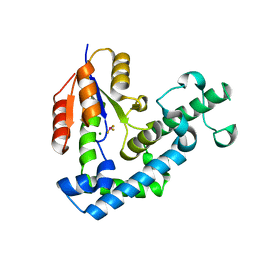 | | Structure of edeya2 complexed with bef3 | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Eyes absent homolog 2 (Drosophila), MAGNESIUM ION | | Authors: | Jung, S.K, Jeong, D.G, Ryu, S.E, Kim, S.J. | | Deposit date: | 2009-05-03 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of ED-Eya2: insight into dual roles as a protein tyrosine phosphatase and a transcription factor
Faseb J., 24, 2010
|
|
2NT2
 
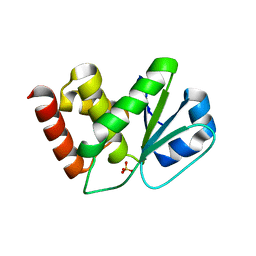 | | Crystal Structure of Slingshot phosphatase 2 | | Descriptor: | Protein phosphatase Slingshot homolog 2, SULFATE ION | | Authors: | Jung, S.K, Jeong, D.G, Yoon, T.S, Kim, J.H, Ryu, S.E, Kim, S.J. | | Deposit date: | 2006-11-06 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human slingshot phosphatase 2.
Proteins, 68, 2007
|
|
2ORY
 
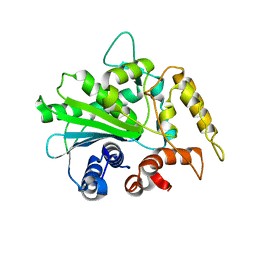 | | Crystal structure of M37 lipase | | Descriptor: | Lipase | | Authors: | Jung, S.K, Jeong, D.G, Kim, S.J. | | Deposit date: | 2007-02-05 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the cold adaptation of psychrophilic M37 lipase from Photobacterium lipolyticum.
Proteins, 71, 2008
|
|
2MXQ
 
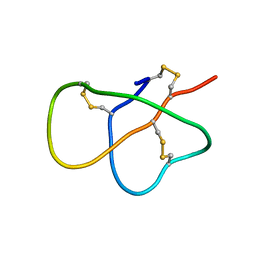 | | The solution structure of DEFA1, a highly potent antimicrobial peptide from the horse | | Descriptor: | Paneth cell-specific alpha-defensin 1 | | Authors: | Jung, S, Michalek, M, Shomali, M, Soennichsen, F.D. | | Deposit date: | 2015-01-12 | | Release date: | 2015-04-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional studies of the highly potent equine antimicrobial peptide DEFA1.
Biochem.Biophys.Res.Commun., 459, 2015
|
|
2LLR
 
 | |
2K35
 
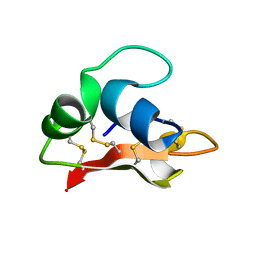 | | Hydramacin-1: Structure and antibacterial activity of a peptide from the basal metazoan Hydra | | Descriptor: | hydramacin-1 | | Authors: | Jung, S, Dingley, A.J, Stanisak, M, Gelhaus, C, Bosch, T, Podschun, R, Leippe, M, Gr tzinger, J. | | Deposit date: | 2008-04-22 | | Release date: | 2008-11-18 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Hydramacin-1, structure and antibacterial activity of a protein from the Basal metazoan hydra.
J.Biol.Chem., 284, 2009
|
|
2LN8
 
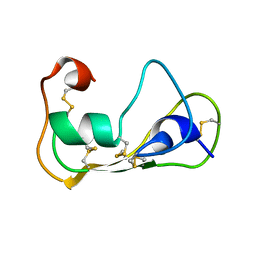 | | The solution structure of theromacin | | Descriptor: | Theromacin | | Authors: | Jung, S, Soennichsen, F.D, Hung, C.-W, Tholey, A, Boidin-Wichlacz, C, Hausgen, W, Gelhaus, C, Desel, C, Podschun, R, Watzig, V, Tasiemski, A, Leippe, M, Groetzinger, J. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Macin family of antimicrobial proteins combines antimicrobial and nerve repair activities.
J.Biol.Chem., 287, 2012
|
|
4ZTT
 
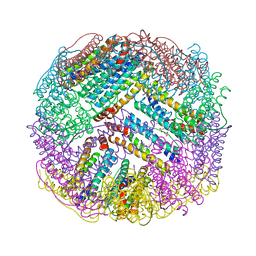 | | Crystal structures of ferritin mutants reveal diferric-peroxo intermediates | | Descriptor: | Bacterial non-heme ferritin, FE (II) ION, FE (III) ION, ... | | Authors: | Kim, S, Park, Y.H, Jung, S.W, Seok, J.H, Chung, Y.B, Lee, D.B, Gowda, G, Lee, J.H, Han, H.R, Cho, A.E, Lee, C, Chung, M.S, Kim, K.H. | | Deposit date: | 2015-05-15 | | Release date: | 2016-06-15 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis of Novel Iron-Uptake Route and Reaction Intermediates in Ferritins from Gram-Negative Bacteria.
J. Mol. Biol., 428, 2016
|
|
7P1J
 
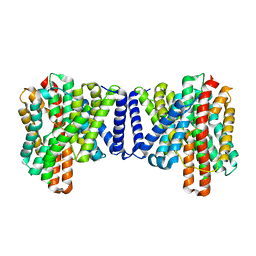 | | Cryo EM structure of bison NHA2 in detergent structure | | Descriptor: | mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P1I
 
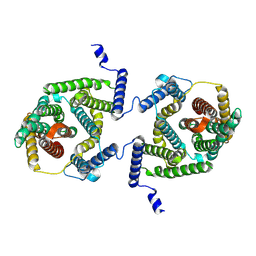 | | Cryo EM structure of bison NHA2 in detergent and N-terminal extension helix | | Descriptor: | mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P1K
 
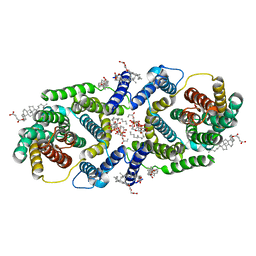 | | Cryo EM structure of bison NHA2 in nano disc structure | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Phosphatidylinositol, mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6Y7G
 
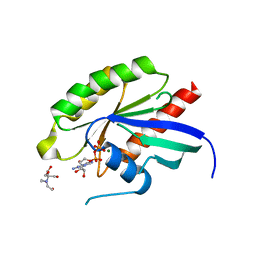 | | Structure of the human RAB3C in complex with GDP | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Diaz-Saez, L, Jung, S, Raux, B, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K.V.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-29 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the human RAB3C in complex with GDP
To Be Published
|
|
6S5H
 
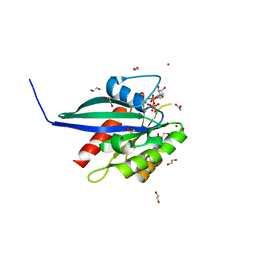 | | Structure of the human RAB38 in complex with GTP | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Diaz-Saez, L, Jung, S, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-07-01 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the human RAB38 in complex with GTP
To Be Published
|
|
3V3V
 
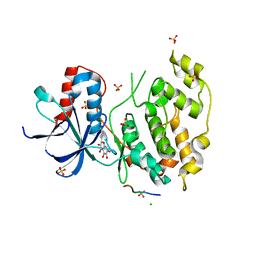 | | Structural and functional analysis of quercetagetin, a natural JNK1 inhibitor | | Descriptor: | 3,5,6,7-TETRAHYDROXY-2-(3,4-DIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, C-Jun-amino-terminal kinase-interacting protein 1, CHLORIDE ION, ... | | Authors: | Baek, S, Kang, N.J, Popowicz, G.M, Arciniega, M, Jung, S.K, Byun, S, Song, N.R, Heo, Y.S, Kim, B.Y, Lee, H.J, Holak, T.A, Augustin, M, Bode, A.M, Huber, R, Dong, Z, Lee, K.W. | | Deposit date: | 2011-12-14 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Analysis of the Natural JNK1 Inhibitor Quercetagetin.
J.Mol.Biol., 425, 2013
|
|
1ZHC
 
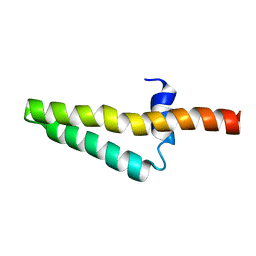 | |
1H8S
 
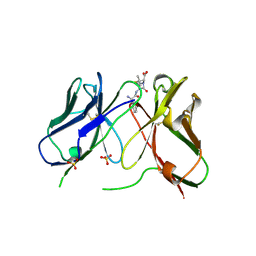 | | Three-dimensional structure of anti-ampicillin single chain Fv fragment complexed with the hapten. | | Descriptor: | (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, MUTANT AL2 6E7P9G, SULFATE ION | | Authors: | Burmester, J, Spinelli, S, Pugliese, L, Krebber, A, Honegger, A, Jung, S, Schimmele, B, Cambillau, C, Pluckthun, A. | | Deposit date: | 2001-02-15 | | Release date: | 2001-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selection, Characterization and X-Ray Structure of Anti-Ampicillin Single-Chain Fv Fragments from Phage-Displayed Murine Antibody Libraries
J.Mol.Biol., 309, 2001
|
|
1H8O
 
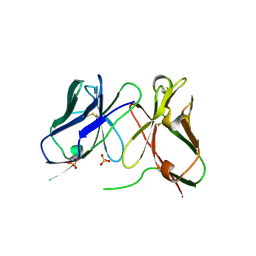 | | Three-dimensional structure of anti-ampicillin single chain Fv fragment. | | Descriptor: | MUTANT AL2 6E7P9G, SULFATE ION | | Authors: | Burmester, J, Spinelli, S, Pugliese, L, Krebber, A, Honegger, A, Jung, S, Schimmele, B, Cambillau, C, Pluckthun, A. | | Deposit date: | 2001-02-14 | | Release date: | 2001-08-02 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Selection, Characterization and X-Ray Structure of Anti-Ampicillin Single-Chain Fv Fragments from Phage-Displayed Murine Antibody Libraries
J.Mol.Biol., 309, 2001
|
|
8JOQ
 
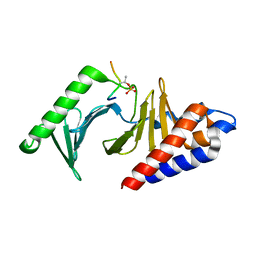 | |
8JOY
 
 | | Plk1 polo-box domain bound to HPV4 L2 residues 251-257 with pThr255 | | Descriptor: | Peptide from Minor capsid protein L2, Serine/threonine-protein kinase PLK1 | | Authors: | Ku, B, Jung, S. | | Deposit date: | 2023-06-09 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structures of Plk1 Polo-Box Domain Bound to the Human Papillomavirus Minor Capsid Protein L2-Derived Peptide.
J.Microbiol, 61, 2023
|
|
6S5F
 
 | | Structure of the human RAB39B in complex with GMPPNP | | Descriptor: | 1,2-ETHANEDIOL, GLYCINE, MAGNESIUM ION, ... | | Authors: | Diaz-Saez, L, Jung, S, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-07-01 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the human RAB39B in complex with GMPPNP
To Be Published
|
|
5C6F
 
 | | Crystal structures of ferritin mutants reveal side-on binding to diiron and end-on cleavage of oxygen | | Descriptor: | Bacterial non-heme ferritin, FE (III) ION, IMIDAZOLE | | Authors: | Kim, S, Kim, K.H, Seok, J.H, Park, Y.H, Jung, S.W, Chung, Y.B, Lee, D.B, Lee, J.H, Han, K.R, Cho, A.E, Lee, C, Chung, M.S. | | Deposit date: | 2015-06-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Novel Iron-Uptake Route and Reaction Intermediates in Ferritins from Gram-Negative Bacteria.
J. Mol. Biol., 428, 2016
|
|
3LJ8
 
 | | Crystal Structure of MKP-4 | | Descriptor: | Tyrosine-protein phosphatase | | Authors: | Jeong, D.G, Yoon, T.S, Jung, S.-K, Park, H.S, Ryu, S.E, Kim, S.J. | | Deposit date: | 2010-01-26 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Exploring binding sites other than the catalytic core in the crystal structure of the catalytic domain of MKP-4
Acta Crystallogr.,Sect.D, 67, 2011
|
|
