7N0H
 
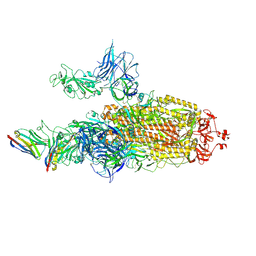 | | CryoEM structure of SARS-CoV-2 spike protein (S-6P, 2-up) in complex with sybodies (Sb45) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jiang, J, Huang, R, Margulies, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7N0G
 
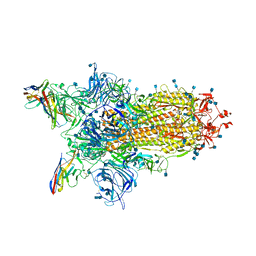 | | CryoEm structure of SARS-CoV-2 spike protein (S-6P, 1-up) in complex with sybodies (Sb45) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jiang, J, Huang, R, Margulies, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
1DVK
 
 | |
5KD7
 
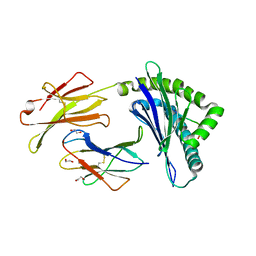 | | Crystal Structure of Murine MHC-I H-2Dd in complex with Murine Beta2-Microglobulin and a Variant of Peptide (PV9) of HIV gp120 MN Isolate (IGPGRAFYV) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Jiang, J, Natarajan, K, Margulies, D. | | Deposit date: | 2016-06-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Effects of Cross-Presentation, Antigen Processing, and Peptide Binding in HIV Evasion of T Cell Immunity.
J. Immunol., 200, 2018
|
|
5KD4
 
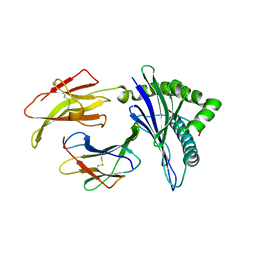 | | Crystal Structure of Murine MHC-I H-2Dd in complex with Murine Beta2-Microglobulin and a Variant of Peptide (PVI10) of HIV gp120 MN Isolate (IGPGRAFYVI) | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-D alpha chain, ... | | Authors: | Jiang, J, Natarajan, K, Margulies, D. | | Deposit date: | 2016-06-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Effects of Cross-Presentation, Antigen Processing, and Peptide Binding in HIV Evasion of T Cell Immunity.
J. Immunol., 200, 2018
|
|
7KGK
 
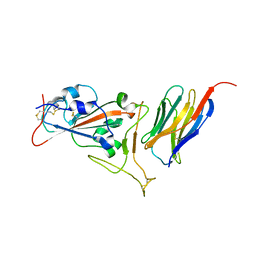 | | Crystal structure of synthetic nanobody (Sb16) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | Sb16, Sybody-16, Synthetic Nanobody, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2020-10-16 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7KLW
 
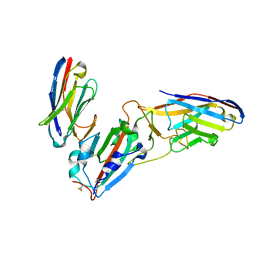 | | Crystal structure of synthetic nanobody (Sb45+Sb68) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | SB45, Synthetic Nanobody, SB68, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2020-11-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7KGJ
 
 | | Crystal structure of synthetic nanobody (Sb45) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | Sb45, Sybody-45, Synthetic Nanobody, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2020-10-16 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7MFV
 
 | | Crystal structure of synthetic nanobody (Sb16) | | Descriptor: | 1,2-ETHANEDIOL, Synthetic Nanobody #16 (Sb16) | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2021-04-11 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
8TQ8
 
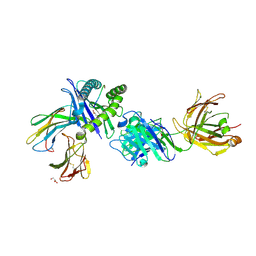 | | Crystal structure of Fab.34.5.8 in complex with MHC-I (H2-Dd) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Fab.34.5.8 Heavy chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQ7
 
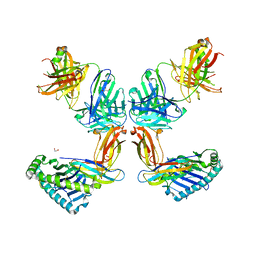 | | Crystal structure of Fab.34.2.12 in complex with MHC-I (H2-Dd) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Fab 34.2.12 Light Chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQ9
 
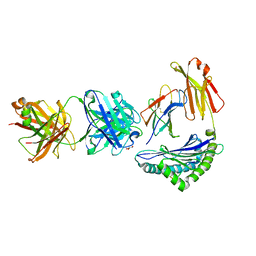 | | Crystal structure of Fab.S19.8 in complex with MHC-I (H2-Dd) | | Descriptor: | Beta-2-microglobulin, Fab.S19.8 Heavy Chain, Fab.S19.8 Light Chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQA
 
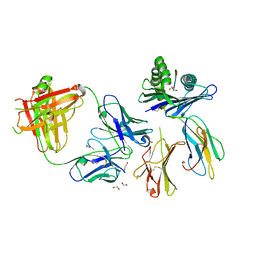 | | Crystal structure of Fab.28.14.8 in complex with MHC-I (H2-Db) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
2NSK
 
 | | Doubled Modified Selenium DNA | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(T4S)P*AP*CP*AP*C)-3' | | Authors: | Jiang, J, Shen, J, Huang, Z. | | Deposit date: | 2006-11-04 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Double Modified Selinium DNA [G(Use)G(Tse)ACAC]
To be Published
|
|
2ESR
 
 | | conserved hypothetical protein- streptococcus pyogenes | | Descriptor: | Methyltransferase, alpha-D-glucopyranose | | Authors: | Jiang, J, Min, T, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-10-26 | | Release date: | 2006-02-07 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of hypothetical protein of Streptococcus Pygenes
To be Published
|
|
6D6V
 
 | | CryoEM structure of Tetrahymena telomerase with telomeric DNA at 4.8 Angstrom resolution | | Descriptor: | DNA (5'-D(P*GP*TP*TP*GP*GP*GP*GP*TP*TP*GP*GP*GP*GP*TP*TP*GP*GP*GP*G)-3'), RNA (159-MER), Telomerase associated protein p65, ... | | Authors: | Jiang, J, Wang, Y, Susac, L, Chan, H, Basu, R, Zhou, Z.H, Feigon, J. | | Deposit date: | 2018-04-22 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of Telomerase with Telomeric DNA.
Cell, 173, 2018
|
|
2DLJ
 
 | |
3BM0
 
 | |
1YUW
 
 | |
7MFU
 
 | | Crystal structure of synthetic nanobody (Sb14+Sb68) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Spike protein S1, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2021-04-11 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
5T7G
 
 | | Crystal Structure of Murine MHC-I H-2Dd in complex with Murine Beta2-Microglobulin and a Variant of Peptide (PT9) of HIV gp120 MN Isolate (IGPGRAFYT) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Jiang, J, Natarajan, K, Margulies, D. | | Deposit date: | 2016-09-04 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Effects of Cross-Presentation, Antigen Processing, and Peptide Binding in HIV Evasion of T Cell Immunity.
J. Immunol., 200, 2018
|
|
2ABM
 
 | | Crystal Structure of Aquaporin Z Tetramer Reveals both Open and Closed Water-conducting Channels | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-O-octyl-beta-D-glucopyranose, ... | | Authors: | Jiang, J, Daniels, B.V, Fu, D. | | Deposit date: | 2005-07-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of AqpZ Tetramer Reveals Two Distinct Arg-189 Conformations Associated with Water Permeation through the Narrowest Constriction of the Water-conducting Channel.
J.Biol.Chem., 281, 2006
|
|
2QWQ
 
 | | Crystal structure of disulfide-bond-crosslinked complex of bovine hsc70 (1-394aa)R171C and bovine Auxilin (810-910aa)D876C in the AMPPNP hydrolyzed form | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Jiang, J, Maes, E.G, Wang, L, Taylor, A.B, Hinck, A.P, Lafer, E.M, Sousa, R. | | Deposit date: | 2007-08-10 | | Release date: | 2007-12-18 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis of J cochaperone binding and regulation of Hsp70.
Mol.Cell, 28, 2007
|
|
2QWL
 
 | | Crystal structure of bovine hsc70 (1-394aa)in the ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Heat shock cognate 71 kDa protein, ... | | Authors: | Jiang, J, Maes, E.G, Wang, L, Taylor, A.B, Hinck, A.P, Lafer, E.M, Sousa, R. | | Deposit date: | 2007-08-10 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of J cochaperone binding and regulation of Hsp70.
Mol.Cell, 28, 2007
|
|
2QW9
 
 | | Crystal structure of bovine hsc70 (1-394aa)in the apo state | | Descriptor: | GLYCEROL, Heat shock cognate 71 kDa protein | | Authors: | Jiang, J, Maes, E.G, Wang, L, Taylor, A.B, Hinck, A.P, Lafer, E.M, Sousa, R. | | Deposit date: | 2007-08-10 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of J cochaperone binding and regulation of Hsp70.
Mol.Cell, 28, 2007
|
|
