1JFZ
 
 | |
6PBZ
 
 | | Crystal structure of Escherichia coli GppA | | Descriptor: | CHLORIDE ION, Guanosine-5'-triphosphate,3'-diphosphate pyrophosphatase | | Authors: | Song, H, Shaw, G.X, Wang, C, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.475 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
6PC3
 
 | | Crystal structure of Helicobacter pylori PPX/GppA in complex with GSP | | Descriptor: | 1,2-ETHANEDIOL, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Song, H, Wang, C, Shaw, G.X, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
6PC2
 
 | | Crystal structure of Helicobacter pylori PPX/GppA in complex with GNP | | Descriptor: | Guanosine pentaphosphate phosphohydrolase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Song, H, Wang, C, Shaw, G.X, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
8SSX
 
 | | Crystal structure of Bacillus anthracis dihydrofolate reductase at 1.65-A resolution | | Descriptor: | 1,2-ETHANEDIOL, Dihydrofolate reductase, SULFATE ION | | Authors: | Shaw, G.X, Li, Y, Wu, Y, Yan, H, Ji, X. | | Deposit date: | 2023-05-09 | | Release date: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Bacillus anthracis dihydrofolate reductase at 1.65-A resolution
To be published
|
|
8SZD
 
 | | Crystal structure of Yersinia pestis dihydrofolate reductase at 1.25-A resolution | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase, MAGNESIUM ION, ... | | Authors: | Shaw, G.X, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2023-05-29 | | Release date: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of Yersinia pestis dihydrofolate reductase at 1.25-A resolution
To be published
|
|
8SZE
 
 | | Crystal structure of Yersinia pestis dihydrofolate reductase in complex with Trimethoprim | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Shaw, G.X, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2023-05-29 | | Release date: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Yersinia pestis dihydrofolate reductase in complex with Trimethoprim
To be published
|
|
1RHH
 
 | | Crystal Structure of the Broadly HIV-1 Neutralizing Fab X5 at 1.90 Angstrom Resolution | | Descriptor: | Fab X5, heavy chain, light chain | | Authors: | Darbha, R, Phogat, S, Labrijn, A.F, Shu, Y, Gu, Y, Andrykovitch, M, Zhang, M.Y, Pantophlet, R, Martin, L, Vita, C, Burton, D.R, Dimitrov, D.S, Ji, X. | | Deposit date: | 2003-11-14 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Broadly Cross-Reactive HIV-1-Neutralizing Fab X5 and Fine Mapping of Its Epitope
Biochemistry, 43, 2004
|
|
1XCA
 
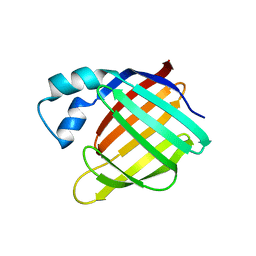 | | APO-CELLULAR RETINOIC ACID BINDING PROTEIN II | | Descriptor: | CELLULAR RETINOIC ACID BINDING PROTEIN TYPE II | | Authors: | Chen, X, Ji, X. | | Deposit date: | 1996-12-31 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of apo-cellular retinoic acid-binding protein type II (R111M) suggests a mechanism of ligand entry.
J.Mol.Biol., 278, 1998
|
|
3R2D
 
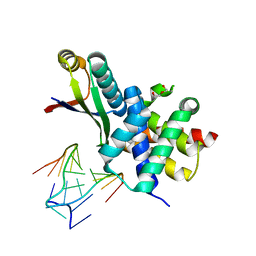 | |
8WDU
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by sucrose density | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
8WDV
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by Ca2+-DEAE | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
5VAG
 
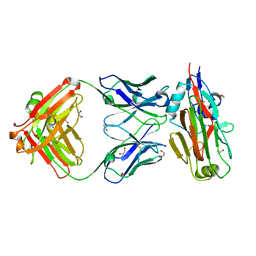 | | Crystal structure of H7-specific antibody m826 in complex with the HA1 domain of hemagglutinin from H7N9 influenza virus | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain of antibody m826, Hemagglutinin, ... | | Authors: | Song, H, Ying, T, Ji, X. | | Deposit date: | 2017-03-26 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Potent Germline-like Human Monoclonal Antibody Targets a pH-Sensitive Epitope on H7N9 Influenza Hemagglutinin.
Cell Host Microbe, 22, 2017
|
|
1TDI
 
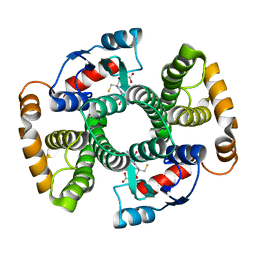 | | Crystal Structure of hGSTA3-3 in Complex with Glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase A3-3 | | Authors: | Gu, Y, Guo, J, Pal, A, Pan, S.S, Zimniak, P, Singh, S.V, Ji, X. | | Deposit date: | 2004-05-22 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human glutathione S-transferase A3-3 and mechanistic implications for its high steroid isomerase activity.
Biochemistry, 43, 2004
|
|
3DJ9
 
 | | Crystal Structure of an isolated, unglycosylated antibody CH2 domain | | Descriptor: | Ig gamma-1 chain C region | | Authors: | Prabakaran, P, Vu, B.K, Gan, J, Dimitrov, D.S, Ji, X. | | Deposit date: | 2008-06-22 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of an isolated unglycosylated antibody C(H)2 domain.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6AP9
 
 | |
5JCU
 
 | |
3H8K
 
 | | Crystal structure of Ube2g2 complxed with the G2BR domain of gp78 at 1.8-A resolution | | Descriptor: | Autocrine motility factor receptor, isoform 2, Ubiquitin-conjugating enzyme E2 G2 | | Authors: | Kalathur, R.C, Das, R, Li, J, Byrd, R.A, Ji, X. | | Deposit date: | 2009-04-29 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78.
Mol.Cell, 34, 2009
|
|
7FIV
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidA and CidBND1-ND2 from wPip(Tunis) | | Descriptor: | CidA_I gamma/2 protein, CidB_I b/2 protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIT
 
 | | Crystal structure of Wolbachia cytoplasmic incompatibility factor CidA from wMel | | Descriptor: | bacteria factor 1 | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIW
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidAwMel(ST) and CidBND1-ND2 from wPip(Pel) | | Descriptor: | ULP_PROTEASE domain-containing protein, bacteria factor 4,CidA I(Zeta/1) protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIU
 
 | | Crystal structure of the DUB domain of Wolbachia cytoplasmic incompatibility factor CidB from wMel | | Descriptor: | ULP_PROTEASE domain-containing protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
5E54
 
 | | Two apo structures of the adenine riboswitch aptamer domain determined using an X-ray free electron laser | | Descriptor: | MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2015-10-07 | | Release date: | 2016-11-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
3ILL
 
 | | Crystal structure of E. coli HPPK(D97A) | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, CHLORIDE ION | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-08-07 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and functional roles of residues D95 and D97 in E. coli HPPK
To be Published
|
|
3ILI
 
 | | Crystal structure of E. coli HPPK(D95A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-08-07 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional roles of residues D95 and D97 in E. coli HPPK
To be Published
|
|
