6MH5
 
 | |
3KAL
 
 | | Structure of homoglutathione synthetase from Glycine max in closed conformation with homoglutathione, ADP, a sulfate ion, and three magnesium ions bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-gamma-glutamyl-L-cysteinyl-beta-alanine, MAGNESIUM ION, ... | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
6E1Q
 
 | | AtGH3.15 acyl acid amido synthetase in complex with 2,4-DB | | Descriptor: | (2,4-DICHLOROPHENOXY)ACETIC ACID, AtGH3.15 acyl acid amido synthetase, PHOSPHATE ION | | Authors: | Sharp, A.M, Lee, S.G, Jez, J.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Modification of auxinic phenoxyalkanoic acid herbicides by the acyl acid amido synthetase GH3.15 from Arabidopsis.
J. Biol. Chem., 293, 2018
|
|
6OMS
 
 | | Arabidopsis GH3.12 with Chorismate | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, 4-substituted benzoates-glutamate ligase GH3.12, ADENOSINE MONOPHOSPHATE | | Authors: | Zubieta, C, Westfall, C.S, Holland, C.K, Jez, J.M. | | Deposit date: | 2019-04-19 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Brassicaceae-specific Gretchen Hagen 3 acyl acid amido synthetases conjugate amino acids to chorismate, a precursor of aromatic amino acids and salicylic acid.
J.Biol.Chem., 294, 2019
|
|
3KAJ
 
 | | Apoenzyme structure of homoglutathione synthetase from Glycine max in open conformation | | Descriptor: | Homoglutathione synthetase | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
3KAK
 
 | | Structure of homoglutathione synthetase from Glycine max in open conformation with gamma-glutamyl-cysteine bound. | | Descriptor: | GAMMA-GLUTAMYLCYSTEINE, Homoglutathione synthetase | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
5J34
 
 | | Isopropylmalate dehydrogenase K232M mutant | | Descriptor: | 3-isopropylmalate dehydrogenase 2, chloroplastic, MAGNESIUM ION, ... | | Authors: | Lee, S.G, Jez, J.M. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.827 Å) | | Cite: | Structure and Mechanism of Isopropylmalate Dehydrogenase from Arabidopsis thaliana: INSIGHTS ON LEUCINE AND ALIPHATIC GLUCOSINOLATE BIOSYNTHESIS.
J.Biol.Chem., 291, 2016
|
|
5WP5
 
 | |
5WP4
 
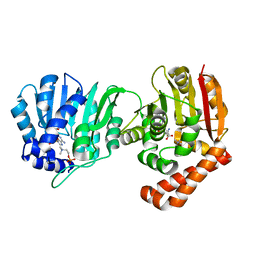 | |
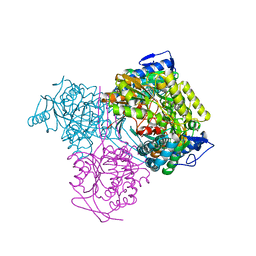
7KQV
 
 | |
6AVH
 
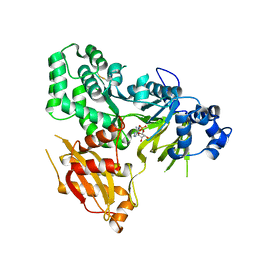 | | GH3.15 acyl acid amido synthetase | | Descriptor: | ADENOSINE MONOPHOSPHATE, GH3.15 acyl acid amido synthetase | | Authors: | Sherp, A.M, Jez, J.M. | | Deposit date: | 2017-09-02 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Arabidopsis thalianaGH3.15 acyl acid amido synthetase has a highly specific substrate preference for the auxin precursor indole-3-butyric acid.
J. Biol. Chem., 293, 2018
|
|
1BQ6
 
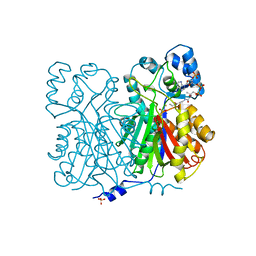 | | CHALCONE SYNTHASE FROM ALFALFA WITH COENZYME A | | Descriptor: | CHALCONE SYNTHASE, COENZYME A, SULFATE ION | | Authors: | Ferrer, J.-L, Bowman, M.E, Jez, J, Dixon, R, Noel, J.P. | | Deposit date: | 1998-08-21 | | Release date: | 1999-08-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1CGK
 
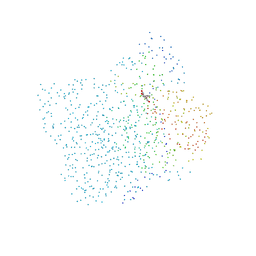 | | CHALCONE SYNTHASE FROM ALFALFA COMPLEXED WITH NARINGENIN | | Descriptor: | NARINGENIN, PROTEIN (CHALCONE SYNTHASE) | | Authors: | Ferrer, J.-L, Jez, J, Bowman, M.E, Dixon, R, Noel, J.P. | | Deposit date: | 1999-03-25 | | Release date: | 1999-09-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1CGZ
 
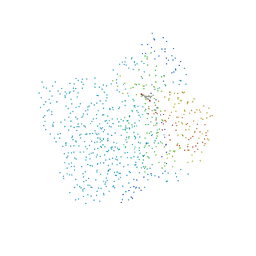 | | CHALCONE SYNTHASE FROM ALFALFA COMPLEXED WITH RESVERATROL | | Descriptor: | PROTEIN (CHALCONE SYNTHASE), RESVERATROL | | Authors: | Ferrer, J.-L, Jez, J, Bowman, M.E, Dixon, R, Noel, J.P. | | Deposit date: | 1999-03-26 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1CHW
 
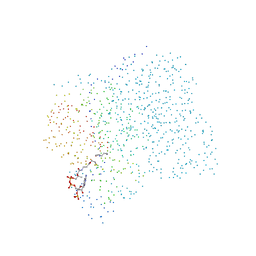 | | CHALCONE SYNTHASE FROM ALFALFA COMPLEXED WITH HEXANOYL-COA | | Descriptor: | HEXANOYL-COENZYME A, PROTEIN (CHALCONE SYNTHASE) | | Authors: | Ferrer, J.-L, Jez, J, Bowman, M.E, Dixon, R, Noel, J.P. | | Deposit date: | 1999-03-30 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1CML
 
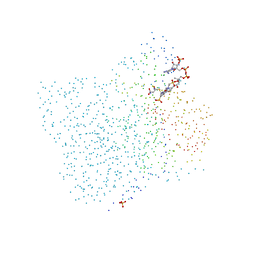 | | CHALCONE SYNTHASE FROM ALFALFA COMPLEXED WITH MALONYL-COA | | Descriptor: | MALONYL-COENZYME A, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), PROTEIN (CHALCONE SYNTHASE), ... | | Authors: | Ferrer, J.-L, Jez, J, Bowman, M.E, Dixon, R, Noel, J.P. | | Deposit date: | 1999-03-30 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1AFS
 
 | | RECOMBINANT RAT LIVER 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE (3-ALPHA-HSD) COMPLEXED WITH NADP AND TESTOSTERONE | | Descriptor: | 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TESTOSTERONE | | Authors: | Bennett, M.J, Albert, R.H, Jez, J.M, Ma, H, Penning, T.M, Lewis, M. | | Deposit date: | 1997-03-13 | | Release date: | 1997-10-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Steroid recognition and regulation of hormone action: crystal structure of testosterone and NADP+ bound to 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase.
Structure, 5, 1997
|
|
6O86
 
 | |
4EQL
 
 | | Crystal Structure of GH3.12 in complex with AMP and salicylate | | Descriptor: | 2-HYDROXYBENZOIC ACID, 4-substituted benzoates-glutamate ligase GH3.12, ADENOSINE MONOPHOSPHATE | | Authors: | Westfall, C, Zubieta, C, Nanao, M, Herrmann, J, Jez, J. | | Deposit date: | 2012-04-19 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for prereceptor modulation of plant hormones by GH3 proteins.
Science, 336, 2012
|
|
4EQ4
 
 | | Crystal structure of seleno-methionine derivatized GH3.12 | | Descriptor: | 2-HYDROXYBENZOIC ACID, 4-substituted benzoates-glutamate ligase GH3.12, ADENOSINE MONOPHOSPHATE | | Authors: | Zubieta, C, Nanao, M, Jez, J, Westfall, C, Kapp, U. | | Deposit date: | 2012-04-18 | | Release date: | 2012-06-20 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.074 Å) | | Cite: | Structural basis for prereceptor modulation of plant hormones by GH3 proteins.
Science, 336, 2012
|
|
4EWV
 
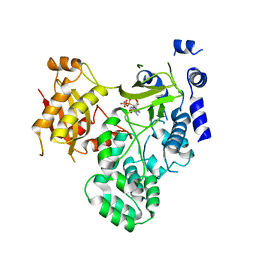 | | Crystal structure of GH3.12 in complex with AMPCPP | | Descriptor: | 4-substituted benzoates-glutamate ligase GH3.12, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MANGANESE (II) ION | | Authors: | Zubieta, C, Nanao, M, Jez, J. | | Deposit date: | 2012-04-28 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | Structural basis for prereceptor modulation of plant hormones by GH3 proteins.
Science, 336, 2012
|
|
4PPU
 
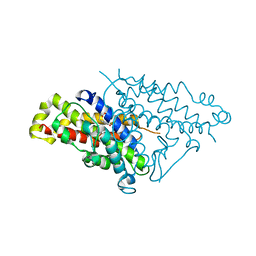 | |
4PPV
 
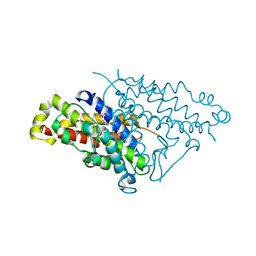 | |
4R6W
 
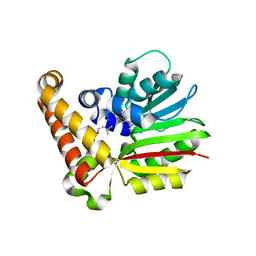 | |
4R6X
 
 | |
