6ETI
 
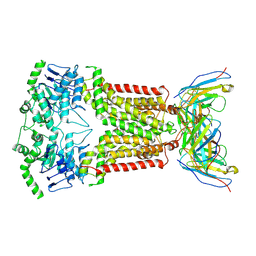 | | Structure of inhibitor-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Jackson, S.M, Manolaridis, I, Kowal, J, Zechner, M, Altmann, K.H, Locher, K.P. | | Deposit date: | 2017-10-26 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of small-molecule inhibition of human multidrug transporter ABCG2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6FFC
 
 | | Structure of an inhibitor-bound ABC transporter | | Descriptor: | ATP-binding cassette sub-family G member 2, ~{tert}-butyl 3-[(2~{S},5~{S},8~{S})-14-cyclopentyloxy-2-(2-methylpropyl)-4,7-bis(oxidanylidene)-3,6,17-triazatetracyclo[8.7.0.0^{3,8}.0^{11,16}]heptadeca-1(10),11,13,15-tetraen-5-yl]propanoate | | Authors: | Jackson, S.M, Manolaridis, I, Kowal, J, Zechner, M, Taylor, N.M.I, Bause, M, Bauer, S, Bartholomaeus, R, Stahlberg, H, Bernhardt, G, Koenig, B, Buschauer, A, Altmann, K.H, Locher, K.P. | | Deposit date: | 2018-01-06 | | Release date: | 2018-04-11 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis of small-molecule inhibition of human multidrug transporter ABCG2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6HIJ
 
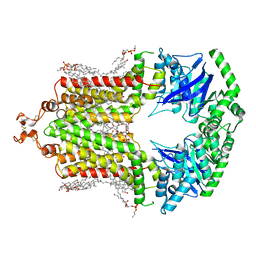 | | Cryo-EM structure of the human ABCG2-MZ29-Fab complex with cholesterol and PE lipids docked | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ATP-binding cassette sub-family G member 2, CHOLESTEROL, ... | | Authors: | Jackson, S.M, Manolaridis, I, Kowal, J, Zechner, M, Taylor, N.M.I, Bause, M, Bauer, S, Bartholomaeus, R, Stahlberg, H, Bernhardt, G, Koenig, B, Buschauer, A, Altmann, K.H, Locher, K.P. | | Deposit date: | 2018-08-30 | | Release date: | 2018-09-19 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis of small-molecule inhibition of human multidrug transporter ABCG2.
Nat.Struct.Mol.Biol., 25, 2018
|
|
6FEQ
 
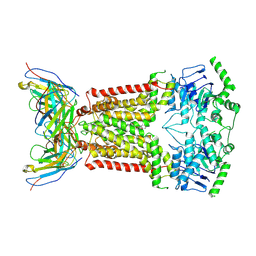 | | Structure of inhibitor-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Jackson, S.M, Manolaridis, I, Kowal, J, Zechner, M, Altmann, K.H, Locher, K.P. | | Deposit date: | 2018-01-03 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of small-molecule inhibition of human multidrug transporter ABCG2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1ZM0
 
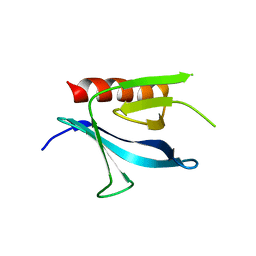 | | Crystal Structure of the Carboxyl Terminal PH Domain of Pleckstrin To 2.1 Angstroms | | Descriptor: | Pleckstrin | | Authors: | Jackson, S.G, Zhang, Y, Zhang, K, Summerfield, R, Haslam, R.J, Junop, M.S. | | Deposit date: | 2005-05-09 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the carboxy-terminal PH domain of pleckstrin at 2.1 Angstroms.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3ISS
 
 | | Crystal structure of enolpyruvyl-UDP-GlcNAc synthase (MurA):UDP-N-acetylmuramic acid:phosphite from Escherichia coli | | Descriptor: | PHOSPHITE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Authors: | Jackson, S.G, Zhang, F, Chindemi, P, Junop, M.S, Berti, P.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence of Kinetic Control of Ligand Binding and Staged Product Release in MurA (Enolpyruvyl UDP-GlcNAc Synthase)-Catalyzed Reactions .
Biochemistry, 48, 2009
|
|
3LS1
 
 | | Crystal Structure of Cyanobacterial PsbQ from Synechocystis sp. PCC 6803 complexed with Zn2+ | | Descriptor: | Sll1638 protein, ZINC ION | | Authors: | Jackson, S.A, Fagerlund, R.D, Wilbanks, S.M, Eaton-Rye, J.J. | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of PsbQ from Synechocystis sp. PCC 6803 at 1.8 A: Implications for Binding and Function in Cyanobacterial Photosystem II
Biochemistry, 49, 2010
|
|
3LS0
 
 | |
2I5F
 
 | | Crystal structure of the C-terminal PH domain of pleckstrin in complex with D-myo-Ins(1,2,3,5,6)P5 | | Descriptor: | (1R,2R,3R,4R,5S,6S)-6-HYDROXYCYCLOHEXANE-1,2,3,4,5-PENTAYL PENTAKIS[DIHYDROGEN (PHOSPHATE)], Pleckstrin | | Authors: | Jackson, S.G, Haslam, R.J, Junop, M.S. | | Deposit date: | 2006-08-24 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis of the carboxy terminal PH domain of pleckstrin bound to D-myo-inositol 1,2,3,5,6-pentakisphosphate.
Bmc Struct.Biol., 7, 2007
|
|
2I5C
 
 | | Crystal structure of the C-terminal PH domain of pleckstrin in complex with D-myo-Ins(1,2,3,4,5)P5 | | Descriptor: | (1R,2S,3R,4S,5S,6R)-6-HYDROXYCYCLOHEXANE-1,2,3,4,5-PENTAYL PENTAKIS[DIHYDROGEN (PHOSPHATE)], Pleckstrin | | Authors: | Jackson, S.G, Haslam, R.J, Junop, M.S. | | Deposit date: | 2006-08-24 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of the carboxy terminal PH domain of pleckstrin bound to D-myo-inositol 1,2,3,5,6-pentakisphosphate.
Bmc Struct.Biol., 7, 2007
|
|
2LNJ
 
 | |
1COA
 
 | | THE EFFECT OF CAVITY CREATING MUTATIONS IN THE HYDROPHOBIC CORE OF CHYMOTRYPSIN INHIBITOR 2 | | Descriptor: | CHYMOTRYPSIN INHIBITOR 2 | | Authors: | Jackson, S.E, Moracci, M, Elmasry, N, Johnson, C.M, Fersht, A.R. | | Deposit date: | 1993-05-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effect of cavity-creating mutations in the hydrophobic core of chymotrypsin inhibitor 2.
Biochemistry, 32, 1993
|
|
8P8A
 
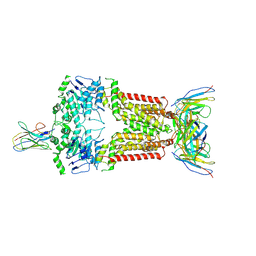 | | Structure of 5D3-Fab and nanobody(Nb17)-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Irobalieva, R.N, Manolaridis, I, Jackson, S.M, Ni, D, Pardon, E, Stahlberg, H, Steyaert, J, Locher, K.P. | | Deposit date: | 2023-05-31 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis of the Allosteric Inhibition of Human ABCG2 by Nanobodies.
J.Mol.Biol., 435, 2023
|
|
8P8J
 
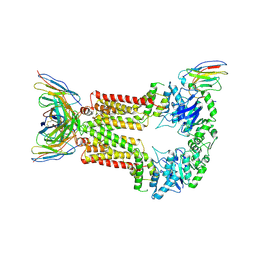 | | Structure of 5D3-Fab and nanobody(Nb96)-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Irobalieva, R.N, Manolaridis, I, Jackson, S.M, Ni, D, Pardon, E, Stahlberg, H, Steyaert, J, Locher, K.P. | | Deposit date: | 2023-06-01 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural Basis of the Allosteric Inhibition of Human ABCG2 by Nanobodies.
J.Mol.Biol., 435, 2023
|
|
8P7W
 
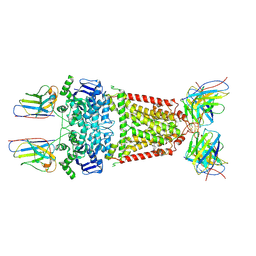 | | Structure of 5D3-Fab and nanobody(Nb8)-bound ABCG2 | | Descriptor: | 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ATP-binding cassette sub-family G member 2, ... | | Authors: | Irobalieva, R.N, Manolaridis, I, Jackson, S.M, Ni, D, Pardon, E, Stahlberg, H, Steyaert, J, Locher, K.P. | | Deposit date: | 2023-05-31 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural Basis of the Allosteric Inhibition of Human ABCG2 by Nanobodies.
J.Mol.Biol., 435, 2023
|
|
1Z56
 
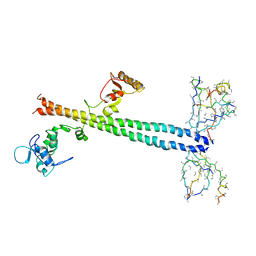 | | Co-Crystal Structure of Lif1p-Lig4p | | Descriptor: | DNA ligase IV, Ligase interacting factor 1 | | Authors: | Dore, A.S, Furnham, N, Davies, O.R, Sibanda, B.L, Chirgadze, D.Y, Jackson, S.P, Pellegrini, L, Blundell, T.L. | | Deposit date: | 2005-03-17 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.92 Å) | | Cite: | Structure of an Xrcc4-DNA ligase IV yeast ortholog complex reveals a novel BRCT interaction mode.
DNA REPAIR, 5, 2006
|
|
5NJG
 
 | | Structure of an ABC transporter: part of the structure that could be built de novo | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3-Fab heavy chain, 5D3-Fab light chain, ... | | Authors: | Taylor, N.M.I, Manolaridis, I, Jackson, S.M, Kowal, J, Stahlberg, H, Locher, K.P. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structure of the human multidrug transporter ABCG2.
Nature, 546, 2017
|
|
5NJ3
 
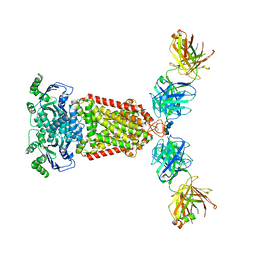 | | Structure of an ABC transporter: complete structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3-Fab heavy chain, 5D3-Fab light chain, ... | | Authors: | Taylor, N.M.I, Manolaridis, I, Jackson, S.M, Kowal, J, Stahlberg, H, Locher, K.P. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structure of the human multidrug transporter ABCG2.
Nature, 546, 2017
|
|
1GXC
 
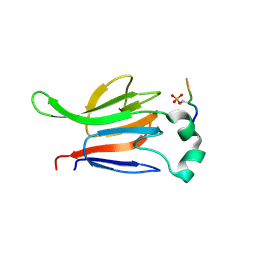 | | FHA domain from human Chk2 kinase in complex with a synthetic phosphopeptide | | Descriptor: | SERINE/THREONINE-PROTEIN KINASE CHK2, SYNTHETIC PHOSPHOPEPTIDE | | Authors: | Li, J, Williams, B.L, Haire, L.F, Goldberg, M, Wilker, E, Durocher, D, Yaffe, M.B, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-13 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Versatility of the Fha Domain in DNA-Damage Signaling by the Tumor Suppressor Kinase Chk2
Mol.Cell, 9, 2002
|
|
6HCO
 
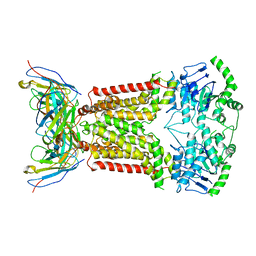 | | Cryo-EM structure of the ABCG2 E211Q mutant bound to estrone 3-sulfate and 5D3-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3-Fab heavy chain, 5D3-Fab light chain, ... | | Authors: | Manolaridis, I, Jackson, S.M, Taylor, N.M.I, Kowal, J, Stahlberg, H, Locher, K.P. | | Deposit date: | 2018-08-15 | | Release date: | 2018-11-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states.
Nature, 563, 2018
|
|
3I0M
 
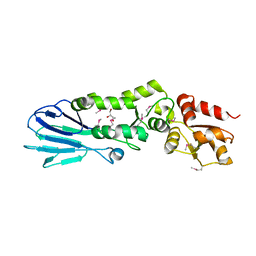 | | Structure of the S. pombe Nbs1 FHA/BRCT-repeat domain | | Descriptor: | DNA repair and telomere maintenance protein nbs1, GLYCEROL | | Authors: | Clapperton, J.A, Lloyd, J, Chapman, J.R, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2009-06-25 | | Release date: | 2009-10-13 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage
Cell(Cambridge,Mass.), 139, 2009
|
|
3I0N
 
 | | Structure of the S. pombe Nbs1 FHA/BRCT-repeat domain | | Descriptor: | DNA repair and telomere maintenance protein nbs1, GLYCEROL | | Authors: | Clapperton, J.A, Lloyd, J, Chapman, J.R, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2009-06-25 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage
Cell(Cambridge,Mass.), 139, 2009
|
|
6HZM
 
 | | Cryo-EM structure of the ABCG2 E211Q mutant bound to ATP and Magnesium (alternative placement of Magnesium into the cryo-EM density) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family G member 2, MAGNESIUM ION | | Authors: | Manolaridis, I, Jackson, S.M, Taylor, N.M.I, Kowal, J, Stahlberg, H, Locher, K.P. | | Deposit date: | 2018-10-23 | | Release date: | 2018-11-14 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states.
Nature, 563, 2018
|
|
6HBU
 
 | | Cryo-EM structure of the ABCG2 E211Q mutant bound to ATP and Magnesium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family G member 2, MAGNESIUM ION | | Authors: | Manolaridis, I, Jackson, S.M, Taylor, N.M.I, Kowal, J, Stahlberg, H, Locher, K.P. | | Deposit date: | 2018-08-13 | | Release date: | 2018-11-07 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states.
Nature, 563, 2018
|
|
3JYZ
 
 | | Crystal structure of Pseudomonas aeruginosa (strain: Pa110594) typeIV pilin in space group P41212 | | Descriptor: | SULFATE ION, Type IV pilin structural subunit | | Authors: | Nguyen, Y, Jackson, S.G, Aidoo, F, Junop, M.S, Burrows, L.L. | | Deposit date: | 2009-09-22 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural characterization of Novel Pseudomonas aeruginosa type IV pilins.
J.Mol.Biol., 395, 2010
|
|
