5A2K
 
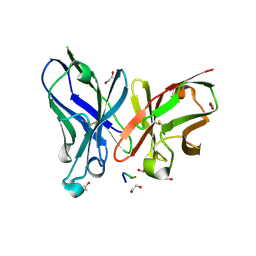 | | Crystal structure of scFv-SM3 in complex with APD-TGalNAc-RP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, ANTIGEN TN, ... | | Authors: | Martinez-Saez, N, Castro-Lopez, J, Valero-Gonzalez, J, Madariaga, D, Companon, I, Somovilla, V.J, Salvado, M, Asensio, J.L, Jimenez-Barbero, J, Avenoza, A, Busto, J.H, Bernardes, G.J.L, Peregrina, J.M, Hurtado-Guerrero, R, Corzana, F. | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Deciphering the Non-Equivalence of Serine and Threonine O-Glycosylation Points: Implications for Molecular Recognition of the Tn Antigen by an Anti-Muc1 Antibody.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5A2J
 
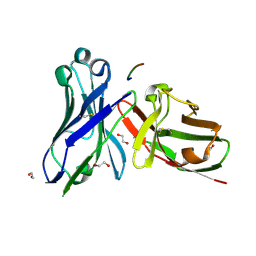 | | Crystal structure of scFv-SM3 in complex with the naked peptide APDTRP | | Descriptor: | 1,2-ETHANEDIOL, SCFV-SM3, THE NAKED PEPTIDE APDTRP | | Authors: | Martinez-Saez, N, Castro-Lopez, J, Valero-Gonzalez, J, Madariaga, D, Companon, I, Somovilla, V.J, Salvado, M, Asensio, J.L, Jimenez-Barbero, J, Avenoza, A, Busto, J.H, Bernardes, G.J.L, Peregrina, J.M, Hurtado-Guerrero, R, Corzana, F. | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Deciphering the Non-Equivalence of Serine and Threonine O-Glycosylation Points: Implications for Molecular Recognition of the Tn Antigen by an Anti-Muc1 Antibody.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5A2I
 
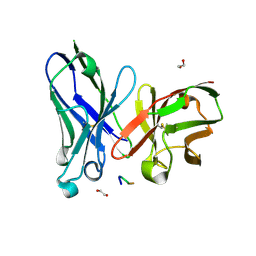 | | Crystal structure of scFv-SM3 in complex with APD-SGalNAc-RP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, ANTIGEN TN, ... | | Authors: | Martinez-Saez, N, Castro-Lopez, J, Valero-Gonzalez, J, Madariaga, D, Companon, I, Somovilla, V.J, Salvado, M, Asensio, J.L, Jimenez-Barbero, J, Avenoza, A, Busto, J.H, Bernardes, G.J.L, Peregrina, J.M, Hurtado-Guerrero, R, Corzana, F. | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Deciphering the Non-Equivalence of Serine and Threonine O-Glycosylation Points: Implications for Molecular Recognition of the Tn Antigen by an Anti-Muc1 Antibody.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5A2L
 
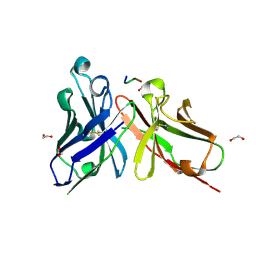 | | Crystal structure of scFv-SM3 in complex with APD-CGalNAc-RP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, MODIFIED ANTIGEN TN, ... | | Authors: | Martinez-Saez, N, Castro-Lopez, J, Valero-Gonzalez, J, Madariaga, D, Companon, I, Somovilla, V.J, Salvado, M, Asensio, J.L, Jimenez-Barbero, J, Avenoza, A, Busto, J.H, Bernardes, G.J.L, Peregrina, J.M, Hurtado-Guerrero, R, Corzana, F. | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Deciphering the Non-Equivalence of Serine and Threonine O-Glycosylation Points: Implications for Molecular Recognition of the Tn Antigen by an Anti-Muc1 Antibody.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3EGS
 
 | | Crystal structure of the HIV-1 broadly neutralizing antibody 2F5 in complex with the gp41 scrambledFP-MPER scrHyb3K construct GIGAFGLLGFLAAGSKK-Ahx-K656NEQELLELDKWASLWN671 soaked in ammonium sulfate | | Descriptor: | 2F5 Fab' heavy chain, 2F5 Fab' light chain, gp41 scrFP-MPER construct | | Authors: | Julien, J.-P, Bryson, S, de la Torre, B.G, Andreu, D, Nieva, J.L, Pai, E.F. | | Deposit date: | 2008-09-11 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural constraints imposed by the conserved fusion peptide on the HIV-1 gp41 epitope recognized by the broadly neutralizing antibody 2F5.
J.Phys.Chem.B, 113, 2009
|
|
3DRT
 
 | | Crystal structure of the HIV-1 broadly neutralizing antibody 2F5 in complex with the gp41 scrambledFP-MPER scrHyb3K construct GIGAFGLLGFLAAGSKK-Ahx-K656NEQELLELDKWASLWN671 | | Descriptor: | 2F5 Fab' heavy chain, 2F5 Fab' light chain, GLYCEROL, ... | | Authors: | Julien, J.-P, Bryson, S, de la Torre, B.G, Andreu, D, Nieva, J.L, Pai, E.F. | | Deposit date: | 2008-07-11 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural constraints imposed by the conserved fusion peptide on the HIV-1 gp41 epitope recognized by the broadly neutralizing antibody 2F5.
J.Phys.Chem.B, 113, 2009
|
|
2ACE
 
 | | NATIVE ACETYLCHOLINESTERASE (E.C. 3.1.1.7) FROM TORPEDO CALIFORNICA | | Descriptor: | ACETYLCHOLINE, ACETYLCHOLINESTERASE | | Authors: | Harel, M, Raves, M.L, Silman, I, Sussman, J.L. | | Deposit date: | 1996-06-23 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of acetylcholinesterase complexed with the nootropic alkaloid, (-)-huperzine A.
Nat.Struct.Biol., 4, 1997
|
|
3CF6
 
 | | Structure of Epac2 in complex with cyclic-AMP and Rap | | Descriptor: | 6-(6-AMINO-PURIN-9-YL)-2-THIOXO-TETRAHYDRO-2-FURO[3,2-D][1,3,2]DIOXAPHOSPHININE-2,7-DIOL, Rap guanine nucleotide exchange factor (GEF) 4, Ras-related protein Rap-1b, ... | | Authors: | Rehmann, H, Arias-Palomo, E, Hadders, M.A, Schwede, F, Llorca, O, Bos, J.L. | | Deposit date: | 2008-03-02 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Epac2 in complex with a cyclic AMP analogue and RAP1B
Nature, 455, 2008
|
|
1ZGC
 
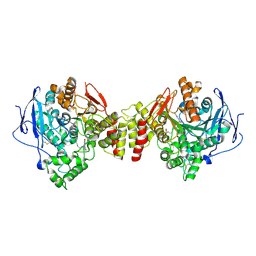 | | Crystal Structure of Torpedo Californica Acetylcholinesterase in Complex With an (RS)-Tacrine(10)-Hupyridone Inhibitor. | | Descriptor: | (5S)-5-{[10-(1,2,3,4-TETRAHYDROACRIDIN-9-YLAMINO)DECYL]AMINO}-5,6,7,8-TETRAHYDROQUINOLIN-2(1H)-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Haviv, H, Wong, D.M, Greenblatt, H.M, Carlier, P.R, Pang, Y.P, Silman, I, Sussman, J.L, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2005-04-21 | | Release date: | 2005-08-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Packing Mediates Enantioselective Ligand Recognition at the Peripheral Site of Acetylcholinesterase
J.Am.Chem.Soc., 127, 2005
|
|
1ZGB
 
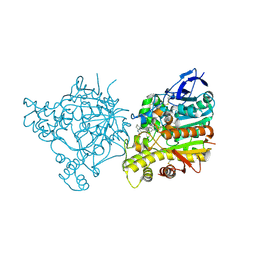 | | Crystal Structure of Torpedo Californica Acetylcholinesterase in Complex With an (R)-Tacrine(10)-Hupyridone Inhibitor. | | Descriptor: | (5R)-5-{[10-(1,2,3,4-TETRAHYDROACRIDIN-9-YLAMINO)DECYL]AMINO}-5,6,7,8-TETRAHYDROQUINOLIN-2(1H)-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Haviv, H, Wong, D.M, Greenblatt, H.M, Carlier, P.R, Pang, Y.P, Silman, I, Sussman, J.L, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2005-04-21 | | Release date: | 2005-08-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Packing Mediates Enantioselective Ligand Recognition at the Peripheral Site of Acetylcholinesterase
J.Am.Chem.Soc., 127, 2005
|
|
1ZNT
 
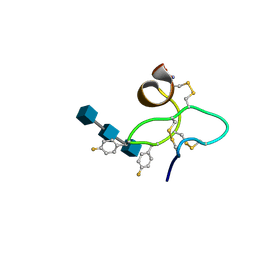 | | 18 NMR structures of AcAMP2-Like Peptide with non Natural Fluoroaromatic Residue (AcAMP2F18Pff/Y20Pff) complex with N,N,N-triacetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMARANTHUS CAUDATUS ANTIMICROBIAL PEPTIDE 2 | | Authors: | Chavez, M.I, Andreu, C, Vidal, P, Aboitiz, N, Freire, F, Groves, P, Asensio, J.L, Asensio, G, Muraki, M, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2005-05-12 | | Release date: | 2005-12-06 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | On the Importance of Carbohydrate-Aromatic Interactions for the Molecular Recognition of Oligosaccharides by Proteins: NMR Studies of the Structure and Binding Affinity of AcAMP2-like Peptides with Non-Natural Naphthyl and Fluoroaromatic Residues
Chemistry, 11, 2005
|
|
1ZUV
 
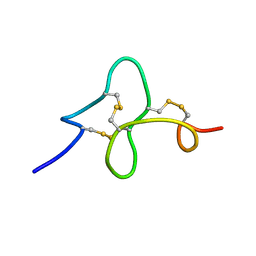 | | 24 NMR structures of AcAMP2-Like Peptide with Phenylalanine 18 mutated to Tryptophan | | Descriptor: | AMARANTHUS CAUDATUS ANTIMICROBIAL PEPTIDE 2 | | Authors: | Chavez, M.I, Andreu, C, Vidal, P, Freire, F, Aboitiz, N, Groves, P, Asensio, J.L, Asensio, G, Muraki, M, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2005-06-01 | | Release date: | 2005-12-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | On the Importance of Carbohydrate-Aromatic Interactions for the Molecular Recognition of Oligosaccharides by Proteins: NMR Studies of the Structure and Binding Affinity of AcAMP2-like Peptides with Non-Natural Naphthyl and Fluoroaromatic Residues
Chemistry, 11, 2005
|
|
1ZWU
 
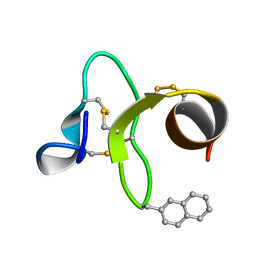 | | 30 NMR structures of AcAMP2-like peptide with non natural beta-(2-naphthyl)-alanine residue. | | Descriptor: | AMARANTHUS CAUDATUS ANTIMICROBIAL PEPTIDE 2 (ACMP2) | | Authors: | Chavez, M.I, Andreu, C, Vidal, P, Freire, F, Aboitiz, N, Groves, P, Asensio, J.L, Asensio, G, Muraki, M, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2005-06-06 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | On the Importance of Carbohydrate-Aromatic Interactions for the Molecular Recognition of Oligosaccharides by Proteins: NMR Studies of the Structure and Binding Affinity of AcAMP2-like Peptides with Non-Natural Naphthyl and Fluoroaromatic Residues.
Chemistry, 11, 2005
|
|
1X7Y
 
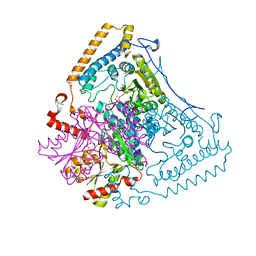 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
1X7Z
 
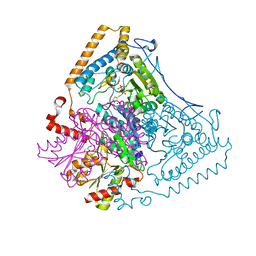 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
1X80
 
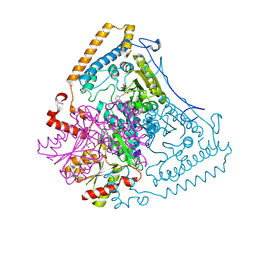 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
1X7W
 
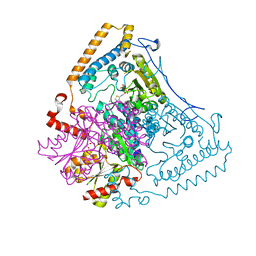 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
1X7X
 
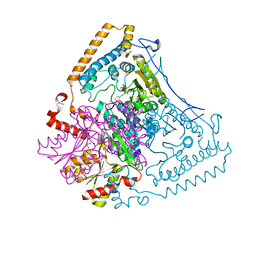 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
1WCI
 
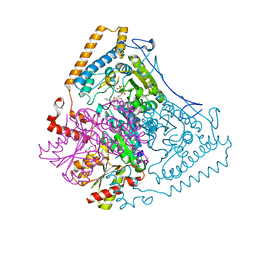 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-3-METHYL-BUTYL-THIAMIN, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-16 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
3G43
 
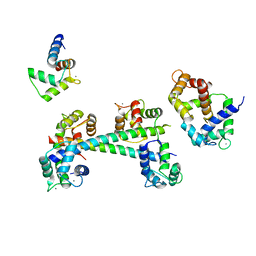 | |
1VRF
 
 | | SOLUTION STRUCTURE OF COMPONENT IV GLYCERA DIBRANCHIATA MONOMERIC HEMOGLOBIN-CO | | Descriptor: | CARBON MONOXIDE, PROTEIN (GLOBIN, MONOMERIC COMPONENT M-IV), ... | | Authors: | Volkman, B.F, Alam, S.L, Satterlee, J.D, Markley, J.L. | | Deposit date: | 1999-03-25 | | Release date: | 1999-04-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of component IV Glycera dibranchiata monomeric hemoglobin-CO.
Biochemistry, 37, 1998
|
|
1ZLI
 
 | | Crystal structure of the tick carboxypeptidase inhibitor in complex with human carboxypeptidase B | | Descriptor: | Carboxypeptidase B, ZINC ION, carboxypeptidase inhibitor | | Authors: | Arolas, J.L, Popowicz, G.M, Lorenzo, J, Sommerhoff, C.P, Huber, R, Aviles, F.X, Holak, T.A. | | Deposit date: | 2005-05-06 | | Release date: | 2005-07-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Three-Dimensional Structures of Tick Carboxypeptidase Inhibitor in Complex with A/B Carboxypeptidases Reveal a Novel Double-headed Binding Mode
J.Mol.Biol., 350, 2005
|
|
1ZFL
 
 | | Solution structure of III-A, the major intermediate in the oxidative folding of leech carboxypeptidase inhibitor | | Descriptor: | Metallocarboxypeptidase inhibitor | | Authors: | Arolas, J.L, D'Silva, L, Popowicz, G.M, Aviles, F.X, Holak, T.A, Ventura, S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-09-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structural characterization and computational predictions of the major intermediate in oxidative folding of leech carboxypeptidase inhibitor
STRUCTURE, 13, 2005
|
|
1ZLH
 
 | | Crystal structure of the tick carboxypeptidase inhibitor in complex with bovine carboxypeptidase A | | Descriptor: | Carboxypeptidase A1, ZINC ION, carboxypeptidase inhibitor | | Authors: | Arolas, J.L, Popowicz, G.M, Lorenzo, J, Sommerhoff, C.P, Huber, R, Aviles, F.X, Holak, T.A. | | Deposit date: | 2005-05-06 | | Release date: | 2005-07-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Three-Dimensional Structures of Tick Carboxypeptidase Inhibitor in Complex with A/B Carboxypeptidases Reveal a Novel Double-headed Binding Mode
J.Mol.Biol., 350, 2005
|
|
3DA6
 
 | | Crystal Structure of human JNK3 complexed with N-(3-methyl-4-(3-(2-(methylamino)pyrimidin-4-yl)pyridin-2-yloxy)naphthalen-1-yl)-1H-benzo[d]imidazol-2-amine | | Descriptor: | Mitogen-activated protein kinase 10, N-[3-methyl-4-({3-[2-(methylamino)pyrimidin-4-yl]pyridin-2-yl}oxy)naphthalen-1-yl]-1H-benzimidazol-2-amine | | Authors: | Cee, V.J, Cheng, A.C, Romero, K, Bellon, S, Mohr, C, Whittington, D.A, Bready, J, Caenepeel, S, Coxon, A, Deak, H.L, Hodous, B.L, Kim, J.L, Lin, J, Nguyen, H, Olivieri, P.R, Patel, V.F, Wang, L, Hughes, P, Geuns-Meyer, S. | | Deposit date: | 2008-05-28 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyridyl-pyrimidine benzimidazole derivatives as potent, selective, and orally bioavailable inhibitors of Tie-2 kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
