3JSS
 
 | |
3JUZ
 
 | |
4A4B
 
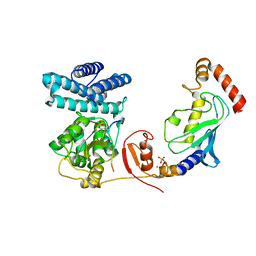 | | Structure of modified phosphoTyr371-c-Cbl-UbcH5B-ZAP-70 complex | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE CBL, TYROSINE-PROTEIN KINASE ZAP-70, ... | | Authors: | Dou, H, Buetow, L, Hock, A, Sibbet, G.J, Vousden, K.H, Huang, D.T. | | Deposit date: | 2011-10-08 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A4C
 
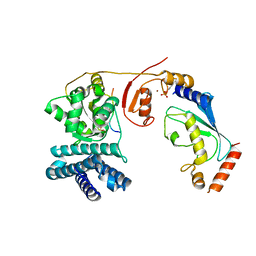 | | Structure of phosphoTyr371-c-Cbl-UbcH5B-ZAP-70 complex | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE CBL, TYROSINE-PROTEIN KINASE ZAP-70, ... | | Authors: | Dou, H, Buetow, L, Hock, A, Sibbet, G.J, Vousden, K.H, Huang, D.T. | | Deposit date: | 2011-10-08 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A49
 
 | | Structure of phosphoTyr371-c-Cbl-UbcH5B complex | | Descriptor: | E3 ubiquitin-protein ligase CBL, POTASSIUM ION, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Dou, H, Buetow, L, Hock, A, Sibbet, G.J, Vousden, K.H, Huang, D.T. | | Deposit date: | 2011-10-07 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.214 Å) | | Cite: | Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl.
Nat. Struct. Mol. Biol., 19, 2012
|
|
5E5B
 
 | |
4BJL
 
 | |
2JBY
 
 | | A viral protein unexpectedly mimics the structure and function of pro- survival Bcl-2 | | Descriptor: | BCL-2 HOMOLOGOUS ANTAGONIST/KILLER 2, M11L PROTEIN, SODIUM ION | | Authors: | Kvansakul, M, Van Delft, M.F, Lee, E.F, Gulbis, J.M, Fairlie, W.D, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2006-12-14 | | Release date: | 2007-03-27 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A structural viral mimic of prosurvival Bcl-2: a pivotal role for sequestering proapoptotic Bax and Bak.
Mol. Cell, 25, 2007
|
|
2MUS
 
 | | HADDOCK calculated model of LIN5001 bound to the HET-s amyloid | | Descriptor: | 3''',4'-bis(carboxymethyl)-2,2':5',2'':5'',2''':5''',2''''-quinquethiophene-5,5''''-dicarboxylic acid, Heterokaryon incompatibility protein s | | Authors: | Hermann, U.S, Schuetz, A.K, Shirani, H, Saban, D, Nuvolone, M, Huang, D.H, Li, B, Ballmer, B, Aslund, A.K.O, Mason, J.J, Rushing, E, Budka, H, Hammarstrom, P, Bockmann, A, Caflisch, A, Meier, B.H, Nilsson, P.K.R, Hornemann, S, Aguzzi, A. | | Deposit date: | 2014-09-16 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-based drug design identifies polythiophenes as antiprion compounds.
Sci Transl Med, 7, 2015
|
|
2JBX
 
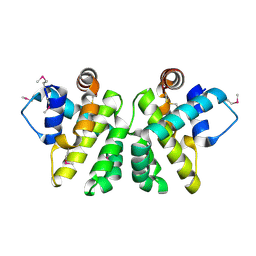 | | Crystal Structure of the myxoma virus anti-apoptotic protein M11L | | Descriptor: | M11L PROTEIN | | Authors: | Kvansakul, M, Van Delft, M.F, Lee, E.F, Gulbis, J.M, Fairlie, W.D, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2006-12-14 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A Structural Viral Mimic of Prosurvival Bcl-2: A Pivotal Role for Sequestering Proapoptotic Bax and Bak.
Mol.Cell, 25, 2007
|
|
2JM6
 
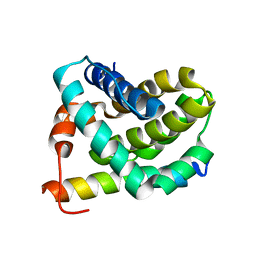 | | Solution structure of MCL-1 complexed with NOXAB | | Descriptor: | Myeloid cell leukemia-1 protein Mcl-1 homolog, Noxa | | Authors: | Czabotar, P.E, Lee, E.F, van Delft, M.F, Day, C.L, Smith, B.J, Huang, D.C.S, Fairlie, W.D, Hinds, M.G, Colman, P.M. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the degradation of Mcl-1 induced by BH3 domains
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2JD5
 
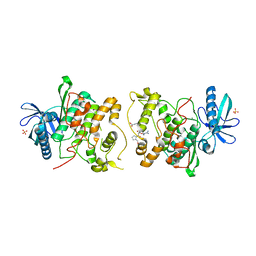 | | Sky1p bound to Npl3p-derived substrate peptide | | Descriptor: | MAGNESIUM ION, NUCLEOLAR PROTEIN 3, SERINE/THREONINE-PROTEIN KINASE SKY1, ... | | Authors: | Nolen, B, Lukasiewicz, R, Adams, J.A, Huang, D, Ghosh, G. | | Deposit date: | 2007-01-04 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Rgg Domain of Npl3P Recruits Sky1P Through Docking Interactions
J.Mol.Biol., 367, 2007
|
|
5ZNE
 
 | |
