5DS1
 
 | | Core domain of the class II small heat-shock protein HSP 17.7 from Pisum sativum | | Descriptor: | 17.1 kDa class II heat shock protein | | Authors: | Hochberg, G.K.A, Laganoswky, A, Allison, T.A, Shepherd, D.A, Benesch, J.L.P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural principles that enable oligomeric small heat-shock protein paralogs to evolve distinct functions.
Science, 359, 2018
|
|
8AN1
 
 | | Structure of a first level Sierpinski triangle formed by a citrate synthase | | Descriptor: | Citrate synthase | | Authors: | Lo, Y.K, Bohn, S, Sendker, F.L, Schuller, J.M, Hochberg, G. | | Deposit date: | 2022-08-04 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Emergence of fractal geometries in the evolution of a metabolic enzyme.
Nature, 628, 2024
|
|
8BEI
 
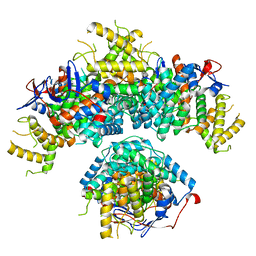 | | Structure of hexameric subcomplexes (Truncation Delta2-6) of the fractal citrate synthase from Synechococcus elongatus PCC7942 | | Descriptor: | Citrate synthase | | Authors: | Lo, Y.K, Bohn, S, Sendker, F.L, Schuller, J.M, Hochberg, G. | | Deposit date: | 2022-10-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Emergence of fractal geometries in the evolution of a metabolic enzyme.
Nature, 628, 2024
|
|
4M5T
 
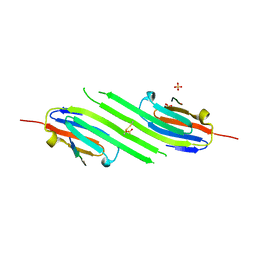 | | Disulfide trapped human alphaB crystallin core domain in complex with C-terminal peptide | | Descriptor: | Alpha-crystallin B chain, SULFATE ION | | Authors: | Laganowsky, A, Cascio, D, Hochberg, G, Sawaya, M.R, Benesch, J.L.P, Robinson, C.V, Eisenberg, D. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structured core domain of alpha B-crystallin can prevent amyloid fibrillation and associated toxicity.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7ZXV
 
 | | Orange Carotenoid Protein Trp-288 BTA mutant | | Descriptor: | CHLORIDE ION, Orange carotenoid-binding protein, beta,beta-caroten-4-one, ... | | Authors: | Moldenhauer, M, Tseng, H.-W, Kraskov, A, Tavraz, N.N, Hildebrandt, P, Hochberg, G, Essen, L.-O, Budisa, N, Korf, L, Maksimov, E.G, Friedrich, T. | | Deposit date: | 2022-05-23 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parameterization of a single H-bond in Orange Carotenoid Protein by atomic mutation reveals principles of evolutionary design of complex chemical photosystems.
Front Mol Biosci, 10, 2023
|
|
8RJL
 
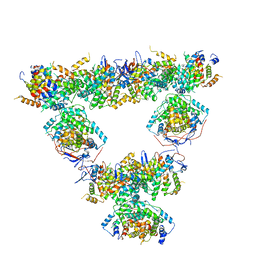 | | Structure of a first order Sierpinski triangle formed by the H369R mutant of the citrate synthase from Synechococcus elongatus | | Descriptor: | Citrate synthase | | Authors: | Lo, Y.K, Bohn, S, Sendker, F.L, Schuller, J.M, Hochberg, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Emergence of fractal geometries in the evolution of a metabolic enzyme.
Nature, 628, 2024
|
|
8RJK
 
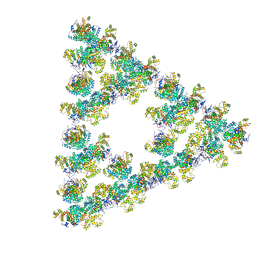 | | Pseudoatomic model of a second-order Sierpinski triangle formed by the citrate synthase from Synechococcus elongatus | | Descriptor: | Citrate synthase | | Authors: | Lo, Y.K, Bohn, S, Sendker, F.L, Schuller, J.M, Hochberg, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.91 Å) | | Cite: | Emergence of fractal geometries in the evolution of a metabolic enzyme.
Nature, 628, 2024
|
|
8QZP
 
 | | Structure of the non-mitochondrial citrate synthase from Ananas comosus | | Descriptor: | Citrate synthase | | Authors: | Lo, Y.K, Bohn, S, Sendker, F.L, Schuller, J.M, Hochberg, G. | | Deposit date: | 2023-10-28 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Frequent transitions in self-assembly across the evolution of a central metabolic enzyme.
Biorxiv, 2024
|
|
8QMW
 
 | | Non-obligately L8S8-complex forming RubisCO derived from ancestral sequence reconstruction and rational engineering in L8S8 complex with substitutions R269W, E271R, L273N | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Layered entrenchment maintains essentiality in protein-protein interactions
Embo J., 2024
|
|
5DS2
 
 | | Core domain of the class I small heat-shock protein HSP 18.1 from Pisum sativum | | Descriptor: | 18.1 kDa class I heat shock protein, SULFATE ION | | Authors: | Shepherd, D.A, Laganowsky, A, Allison, T.M, Hochberg, G.K.A, Benesch, J.L.P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural principles that enable oligomeric small heat-shock protein paralogs to evolve distinct functions.
Science, 359, 2018
|
|
8QMV
 
 | | L2 forming RubisCO derived from ancestral sequence reconstruction of the last common ancestor of Form I'' and Form I RubisCOs | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Layered entrenchment maintains essentiality in the evolution of Form I Rubisco complexes
Biorxiv, 2024
|
|
7QSX
 
 | | Non-obligately L8S8-complex forming RubisCO derived from ancestral sequence reconstruction and rational engineering in L8 complex | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
7QT1
 
 | | Non-obligately L8S8-complex forming RubisCO derived from ancestral sequence reconstruction and rational engineering in L8S8 complex with substitution e170N | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit, ... | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
7QSW
 
 | | L8S8-complex forming RubisCO derived from ancestral sequence reconstruction of the last common ancestor of SSU-bearing Form I RubisCOs | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit, ... | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
7QVI
 
 | | Fiber-forming RubisCO derived from ancestral sequence reconstruction and rational engineering | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit | | Authors: | Schulz, L, Zarzycki, J, Prinz, S, Schuller, J.M, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
7QSV
 
 | | L8-complex forming RubisCO derived from ancestral sequence reconstruction of the last common ancestor of Form I'' and Form I RubisCOs | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
7QSY
 
 | | Non-obligately L8S8-complex forming RubisCO derived from ancestral sequence reconstruction and rational engineering in L8S8 complex | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit, ... | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
7QSZ
 
 | | Non-obligately L8S8-complex forming RubisCO derived from ancestral sequence reconstruction and rational engineering in L8 complex with substitution e170N | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
8QWB
 
 | |
8BW1
 
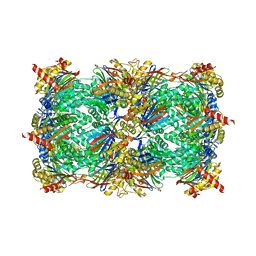 | | Yeast 20S proteasome in complex with an engineered fellutamide derivative (C14QAL) | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bozhueyuek, K.A.J, Praeve, L, Kegler, C, Kaiser, S, Shi, Y, Kuttenlochner, W, Schenk, L, Groll, M, Hochberg, G.K.A, Bode, H.B. | | Deposit date: | 2022-12-06 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Evolution-inspired engineering of nonribosomal peptide synthetases.
Science, 383, 2024
|
|
8C0Z
 
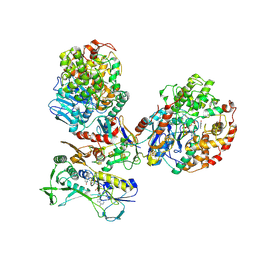 | | CryoEM structure of a tungsten-containing aldehyde oxidoreductase from Aromatoleum aromaticum | | Descriptor: | Aldehyde:ferredoxin oxidoreductase,tungsten-containing, BENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Winiarska, A, Ramirez-Amador, F, Hege, D, Gemmecker, Y, Prinz, S, Hochberg, G, Heider, J, Szaleniec, M, Schuller, J.M. | | Deposit date: | 2022-12-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | A bacterial tungsten-containing aldehyde oxidoreductase forms an enzymatic decorated protein nanowire.
Sci Adv, 9, 2023
|
|
