966C
 
 | | CRYSTAL STRUCTURE OF FIBROBLAST COLLAGENASE-1 COMPLEXED TO A DIPHENYL-ETHER SULPHONE BASED HYDROXAMIC ACID | | Descriptor: | CALCIUM ION, MMP-1, N-HYDROXY-2-[4-(4-PHENOXY-BENZENESULFONYL)-TETRAHYDRO-PYRAN-4-YL]-ACETAMIDE, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-07 | | Release date: | 1999-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
830C
 
 | | COLLAGENASE-3 (MMP-13) COMPLEXED TO A SULPHONE-BASED HYDROXAMIC ACID | | Descriptor: | 4-[4-(4-CHLORO-PHENOXY)-BENZENESULFONYLMETHYL]-TETRAHYDRO-PYRAN-4-CARBOXYLIC ACID HYDROXYAMIDE, CALCIUM ION, MMP-13, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-06 | | Release date: | 1999-08-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
456C
 
 | | CRYSTAL STRUCTURE OF COLLAGENASE-3 (MMP-13) COMPLEXED TO A DIPHENYL-ETHER SULPHONE BASED HYDROXAMIC ACID | | Descriptor: | 2-{4-[4-(4-CHLORO-PHENOXY)-BENZENESULFONYL]-TETRAHYDRO-PYRAN-4-YL}-N-HYDROXY-ACETAMIDE, CALCIUM ION, MMP-13, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-06 | | Release date: | 1999-08-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
3LW7
 
 | | The Crystal Structure of an Adenylate kinase-related protein bound to AMP from sulfolobus solfataricus to 2.3A | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenylate kinase related protein (AdkA-like) | | Authors: | Stein, A.J, Sather, A, Hendricks, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-23 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of an Adenylate kinase-related protein bound to AMP from sulfolobus solfataricus to 2.3A
To be Published
|
|
3LWA
 
 | | The Crystal Structure of a Secreted Thiol-disulfide Isomerase from Corynebacterium glutamicum to 1.75A | | Descriptor: | CALCIUM ION, Secreted thiol-disulfide isomerase | | Authors: | Stein, A.J, Weger, A, Hendricks, R, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-23 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of a Secreted Thiol-disulfide Isomerase from Corynebacterium glutamicum to 1.75A
To be Published
|
|
3D1L
 
 | | Crystal structure of putative NADP oxidoreductase BF3122 from Bacteroides fragilis | | Descriptor: | 2-MERCAPTO-PROPION ALDEHYDE, CHLORIDE ION, Putative NADP oxidoreductase BF3122 | | Authors: | Chang, C, Hendricks, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of putative NADP oxidoreductase BF3122 from Bacteroides fragilis.
To be Published
|
|
3DNS
 
 | |
3EAG
 
 | |
3CAN
 
 | | Crystal structure of a domain of pyruvate-formate lyase-activating enzyme from Bacteroides vulgatus ATCC 8482 | | Descriptor: | Pyruvate-formate lyase-activating enzyme | | Authors: | Nocek, B, Hendricks, R, Hatzos, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a domain of pyruvate-formate lyase-activating enzyme from Bacteroides vulgatus ATCC 8482.
To be Published
|
|
3CO5
 
 | | Crystal structure of sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae | | Descriptor: | BETA-MERCAPTOETHANOL, Putative two-component system transcriptional response regulator | | Authors: | Osipiuk, J, Hendricks, R, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of Sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae.
To be Published
|
|
3C8M
 
 | |
3FDJ
 
 | | The structure of a DegV family protein from Eubacterium eligens. | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DegV family protein, ... | | Authors: | Cuff, M.E, Hendricks, R, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-25 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a DegV family protein from Eubacterium eligens.
TO BE PUBLISHED
|
|
3G1C
 
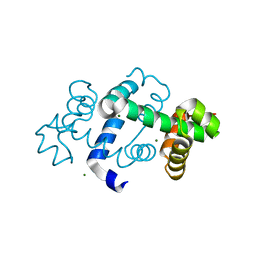 | | The crystal structure of a TrpR like protein from Eubacterium eligens ATCC 27750 | | Descriptor: | MAGNESIUM ION, The TrpR like protein from Eubacterium eligens ATCC 27750 | | Authors: | Zhang, R, Hendricks, R, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-29 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a TrpR like protein from Eubacterium eligens ATCC 27750
To be Published
|
|
3HE0
 
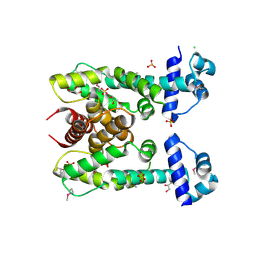 | | The Structure of a Putative Transcriptional Regulator TetR Family Protein from Vibrio parahaemolyticus. | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Cuff, M.E, Hendricks, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-07 | | Release date: | 2009-07-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of a Putative Transcriptional Regulator TetR Family Protein from Vibrio parahaemolyticus.
TO BE PUBLISHED
|
|
3HA9
 
 | | The 1.7A Crystal Structure of a Thioredoxin-like Protein from Aeropyrum pernix | | Descriptor: | uncharacterized Thioredoxin-like protein | | Authors: | Stein, A.J, Cuff, M.E, Sather, A, Hendricks, R, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-01 | | Release date: | 2009-05-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7A Crystal Structure of a Thioredoxin-like Protein from Aeropyrum pernix
To be Published
|
|
3IG2
 
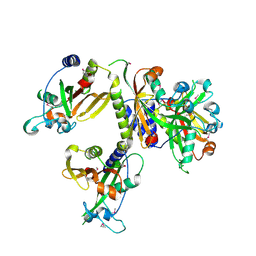 | | The Crystal Structure of a Putative Phenylalanyl-tRNA synthetase (PheRS) beta chain domain from Bacteroides fragilis to 2.1A | | Descriptor: | MAGNESIUM ION, Phenylalanyl-tRNA synthetase beta chain | | Authors: | Stein, A.J, Sather, A, Hendricks, R, Keigher, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-27 | | Release date: | 2009-09-01 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Crystal Structure of a Putative Phenylalanyl-tRNA synthetase (PheRS) beta chain domain from Bacteroides fragilis to 2.1A
To be Published
|
|
3JSA
 
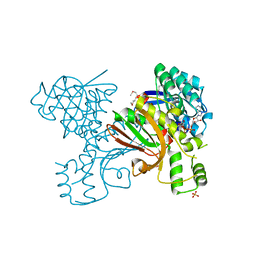 | | Homoserine dehydrogenase from Thermoplasma volcanium complexed with NAD | | Descriptor: | Homoserine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Osipiuk, J, Nocek, B, Hendricks, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-09 | | Release date: | 2009-09-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structure of Homoserine dehydrogenase from Thermoplasma volcanium
To be Published
|
|
3LLV
 
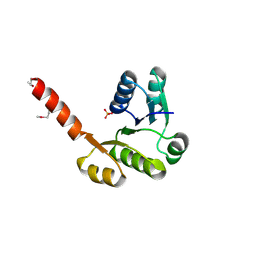 | | The Crystal Structure of the NAD(P)-binding domain of an Exopolyphosphatase-related protein from Archaeoglobus fulgidus to 1.7A | | Descriptor: | Exopolyphosphatase-related protein, PHOSPHATE ION | | Authors: | Stein, A.J, Chang, C, Weger, A, Hendricks, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the NAD(P)-binding domain of an Exopolyphosphatase-related protein from Archaeoglobus fulgidus to 1.7A
To be Published
|
|
3HNW
 
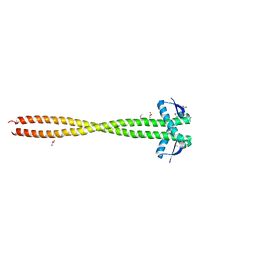 | | Crystal Structure of a Basic Coiled-Coil Protein of Unknown Function from Eubacterium eligens ATCC 27750 | | Descriptor: | GLYCEROL, IODIDE ION, uncharacterized protein | | Authors: | Kim, Y, Hendricks, R, Keigher, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-06-01 | | Release date: | 2009-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Crystal Structure of a Basic Coiled-Coil Protein of Unknown Function from Eubacterium eligens ATCC 27750
To be Published
|
|
