4RV0
 
 | | Crystal structure of TN complex | | Descriptor: | Nuclear protein localization protein 4 homolog, SULFATE ION, Transitional endoplasmic reticulum ATPase TER94 | | Authors: | Hao, Q, Jiao, S, Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2014-11-23 | | Release date: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of TN complex
To be Published
|
|
3NS2
 
 | | High-resolution structure of pyrabactin-bound PYL2 | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2 | | Authors: | Hao, Q, Yin, P, Yan, C, Yuan, X, Wang, J, Yan, N. | | Deposit date: | 2010-07-01 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.634 Å) | | Cite: | Single amino acid alteration between Valine and Isoleucine determines the distinct pyrabactin selectivity by PYL1 and PYL2
J.Biol.Chem., 285, 2010
|
|
4O27
 
 | | Crystal structure of MST3-MO25 complex with WIF motif | | Descriptor: | 5-mer peptide from serine/threonine-protein kinase 24, ADENOSINE-5'-DIPHOSPHATE, Calcium-binding protein 39, ... | | Authors: | Hao, Q, Feng, M, Zhou, Z.C. | | Deposit date: | 2013-12-16 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.185 Å) | | Cite: | Structural insights into regulatory mechanisms of MO25-mediated kinase activation.
J.Struct.Biol., 186, 2014
|
|
5F21
 
 | | human CD38 in complex with nanobody MU375 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Hao, Q, Zhang, H. | | Deposit date: | 2015-12-01 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
3RT0
 
 | | Crystal structure of PYL10-HAB1 complex in the absence of abscisic acid (ABA) | | Descriptor: | Abscisic acid receptor PYL10, MAGNESIUM ION, Protein phosphatase 2C 16 | | Authors: | Hao, Q, Yin, P, Li, W, Wang, L, Yan, C, Wang, J, Yan, N. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | The Molecular Basis of ABA-Independent Inhibition of PP2Cs by a Subclass of PYL Proteins
Mol.Cell, 42, 2011
|
|
3RT2
 
 | | Crystal structure of apo-PYL10 | | Descriptor: | Abscisic acid receptor PYL10 | | Authors: | Hao, Q, Yin, P, Li, W, Wang, L, Yan, C, Wang, J, Yan, N. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Molecular Basis of ABA-Independent Inhibition of PP2Cs by a Subclass of PYL Proteins
Mol.Cell, 42, 2011
|
|
5FB7
 
 | |
3BNU
 
 | | Crystal structure of polyamine oxidase FMS1 from Saccharomyces cerevisiae in complex with bis-(3S,3'S)-methylated spermine | | Descriptor: | (3S,3'S)-N~1~,N~1~'-butane-1,4-diyldibutane-1,3-diamine, FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1 | | Authors: | Huang, Q, Hao, Q. | | Deposit date: | 2007-12-14 | | Release date: | 2008-01-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the substrate stereospecificity of FMS1.
To Be Published
|
|
3U4I
 
 | | CD38 structure-based inhibitor design using the N1-cyclic inosine 5'-diphosphate ribose template | | Descriptor: | ADP-ribosyl cyclase 1, Cyclic adenosine 5'-diphosphocarbocyclic ribose | | Authors: | Liu, Q, Hao, Q, Lee, H.C, Graeff, R. | | Deposit date: | 2011-10-08 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.118 Å) | | Cite: | CD38 Structure-Based Inhibitor Design Using the N1-Cyclic Inosine 5'-Diphosphate Ribose Template
Plos One, 8, 2013
|
|
3U31
 
 | |
5J5K
 
 | | CRYSTAL STRUCTURE OF AFMP4P IN COMPLEX WITH PALMITIC ACID | | Descriptor: | PALMITIC ACID, Uncharacterized protein, alpha-D-mannopyranose | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2016-04-03 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Novel Class Of Virulence Factors In Penicillium Marneffei And Aspergillus Fumigatus Enhances Intracellular Survival In Monocytes By Arachidonic Acid Binding
To Be Published
|
|
2B5H
 
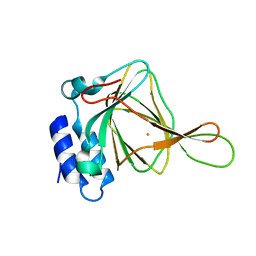 | | 1.5 A Resolution Crystal Structure of Recombinant R. Norvegicus Cysteine Dioxygenase | | Descriptor: | Cysteine dioxygenase type I, FE (III) ION | | Authors: | Simmons, C.R, Liu, Q, Huang, Q, Hao, Q, Begley, T.P, Karplus, P.A, Stipanuk, M.H. | | Deposit date: | 2005-09-28 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Mammalian Cysteine Dioxygenase: A NOVEL MONONUCLEAR IRON CENTER FOR CYSTEINE THIOL OXIDATION.
J.Biol.Chem., 281, 2006
|
|
3BI5
 
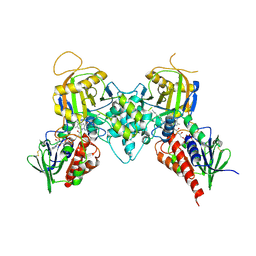 | |
3BI4
 
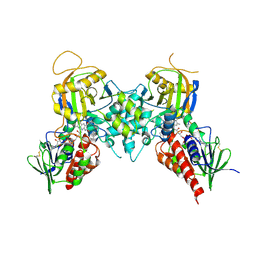 | | Crystal structures of fms1 in complex with its inhibitors | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[(1E,2Z)-but-2-en-1-ylidene]-N'-[(2E)-but-2-en-1-ylidene]butane-1,4-diamine, Polyamine oxidase FMS1 | | Authors: | Huang, Q, Hao, Q. | | Deposit date: | 2007-11-29 | | Release date: | 2008-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of fms1 in complex with its inhibitors
To be Published
|
|
3BI2
 
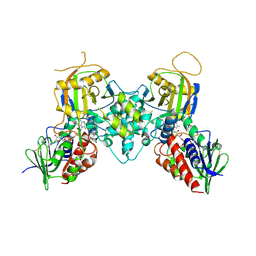 | |
3BNM
 
 | | Crystal structure of polyamine oxidase FMS1 from Saccharomyces cerevisiae in complex with bis-(3R,3'R)-methylated spermine | | Descriptor: | (3R,3'R)-N~1~,N~1~'-butane-1,4-diyldibutane-1,3-diamine, FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1 | | Authors: | Huang, Q, Hao, Q. | | Deposit date: | 2007-12-14 | | Release date: | 2008-01-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the substrate stereospecificity of FMS1.
To Be Published
|
|
1Z6L
 
 | |
3KDI
 
 | | Structure of (+)-ABA bound PYL2 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Putative uncharacterized protein At2g26040 | | Authors: | Yin, P, Fan, H, Hao, Q, Yuan, X, Yan, N. | | Deposit date: | 2009-10-22 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.379 Å) | | Cite: | Structural insights into the mechanism of abscisic acid signaling by PYL proteins
Nat.Struct.Mol.Biol., 16, 2009
|
|
1QWO
 
 | | Crystal structure of a phosphorylated phytase from Aspergillus fumigatus, revealing the structural basis for its heat resilience and catalytic pathway | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, phytase | | Authors: | Xiang, T, Liu, Q, Deacon, A.M, Koshy, M, Kriksunov, I.A, Lei, X.G, Hao, Q, Thiel, D.J. | | Deposit date: | 2003-09-03 | | Release date: | 2004-06-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Heat-resilient Phytase from Aspergillus fumigatus, Carrying a Phosphorylated Histidine
J.Mol.Biol., 339, 2004
|
|
1TW9
 
 | | Glutathione Transferase-2, apo form, from the nematode Heligmosomoides polygyrus | | Descriptor: | glutathione S-transferase 2 | | Authors: | Schuller, D.J, Liu, Q, Kriksunov, I.A, Campbell, A.M, Barrett, J, Brophy, P.M, Hao, Q. | | Deposit date: | 2004-06-30 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of a new class of glutathione transferase from the model human hookworm nematode Heligmosomoides polygyrus.
Proteins, 61, 2005
|
|
1XPQ
 
 | | Crystal structure of fms1, a polyamine oxidase from yeast | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1, SPERMINE | | Authors: | Huang, Q, Liu, Q, Hao, Q. | | Deposit date: | 2004-10-09 | | Release date: | 2005-04-26 | | Last modified: | 2021-12-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structures of Fms1 and its complex with spermine reveal substrate specificity.
J.Mol.Biol., 348, 2005
|
|
1YY5
 
 | |
1A8Z
 
 | | STRUCTURE DETERMINATION OF A 16.8KDA COPPER PROTEIN RUSTICYANIN AT 2.1A RESOLUTION USING ANOMALOUS SCATTERING DATA WITH DIRECT METHODS | | Descriptor: | COPPER (I) ION, RUSTICYANIN | | Authors: | Harvey, I, Hao, Q, Duke, E.M.H, Ingledew, W.J, Hasnain, S.S. | | Deposit date: | 1998-03-30 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure determination of a 16.8 kDa copper protein at 2.1 A resolution using anomalous scattering data with direct methods.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2YPN
 
 | | Hydroxymethylbilane synthase | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, PROTEIN (HYDROXYMETHYLBILANE SYNTHASE) | | Authors: | Nieh, Y.P, Raftery, J, Weisgerber, S, Habash, J, Schotte, F, Ursby, T, Wulff, M, Haedener, A, Campbell, J.W, Hao, Q, Helliwell, J.R. | | Deposit date: | 1999-01-11 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Accurate and highly complete synchrotron protein crystal Laue diffraction data using the ESRF CCD and the Daresbury Laue software
J.Synchrotron Radiat., 6, 1999
|
|
4F4U
 
 | |
