7AD2
 
 | |
8AV5
 
 | |
7ANT
 
 | | Structure of CYP153A from Polaromonas sp. | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450, HEME C | | Authors: | Zukic, E, Rowlinson, B, Sharma, M, Hoffman, S, Hauer, B, Grogan, G. | | Deposit date: | 2020-10-12 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
Acs Catalysis, 11, 2021
|
|
7AO7
 
 | | Structure of CYP153A from Polaromonas sp. in complex with octan-1-ol | | Descriptor: | Cytochrome P450, OCTAN-1-OL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zukic, E, Rowlinson, B, Sharma, M, Hoffmann, S, Hauer, B, Grogan, G. | | Deposit date: | 2020-10-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
Acs Catalysis, 11, 2021
|
|
8BJ5
 
 | |
8CI9
 
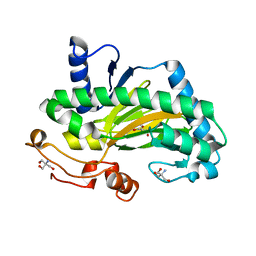 | | Deoxypodophyllotoxin Synthase in complex with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxypodophyllotoxin synthase, FE (III) ION | | Authors: | Ingold, Z, Lichman, B, Grogan, G. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure and mutation of deoxypodophyllotoxin synthase (DPS) from Podophyllum hexandrum
Front Catal, 3, 2023
|
|
5M46
 
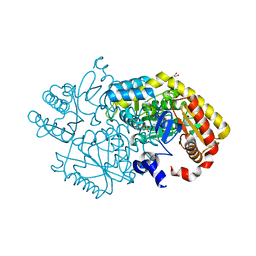 | | Alpha-amino epsilon-caprolactam racemase (ACLR) from Rhizobacterium freirei | | Descriptor: | 1,2-ETHANEDIOL, Aminotransferase class-III, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Frese, A, Sutton, P.W, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-10-18 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Snapshots of the Catalytic Cycle of the Industrial Enzyme alpha-Amino-epsilon-Caprolactam Racemase (ACLR) Observed Using X-ray Crystallography
Acs Catalysis, 7, 2017
|
|
5M4D
 
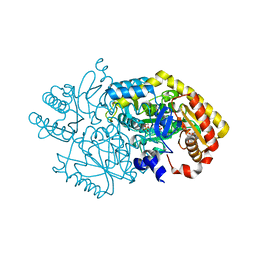 | | Alpha-amino epsilon-caprolactam racemase K241A mutant in complex with D-ACL (external aldimine) | | Descriptor: | 1,2-ETHANEDIOL, Aminotransferase class-III, [6-methyl-5-oxidanyl-4-[(~{E})-[(3~{R})-2-oxidanylideneazepan-3-yl]iminomethyl]pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Frese, A, Sutton, P.W, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-10-18 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Snapshots of the Catalytic Cycle of the Industrial Enzyme alpha-Amino-epsilon-Caprolactam Racemase (ACLR) Observed Using X-ray Crystallography
Acs Catalysis, 7, 2017
|
|
5M49
 
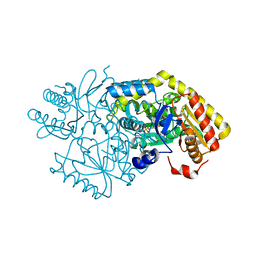 | | Alpha-amino epsilon-caprolactam racemase in complex with PLP and D/L alpha amino epsilon-caprolactam (internal aldimine) | | Descriptor: | (3~{R})-3-azanylazepan-2-one, (3~{S})-3-azanylazepan-2-one, Aminotransferase class-III, ... | | Authors: | Frese, A, Sutton, P.W, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-10-18 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Snapshots of the Catalytic Cycle of the Industrial Enzyme alpha-Amino-epsilon-Caprolactam Racemase (ACLR) Observed Using X-ray Crystallography
Acs Catalysis, 7, 2017
|
|
5M4B
 
 | | Alpha-amino epsilon-caprolactam racemase D210A mutant in complex with PLP and geminal diamine intermediate | | Descriptor: | 1,2-ETHANEDIOL, Aminotransferase class-III, [6-methyl-5-oxidanyl-4-[(~{E})-[(3~{R})-2-oxidanylideneazepan-3-yl]iminomethyl]pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Frese, A, Sutton, P.W, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-10-18 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Snapshots of the Catalytic Cycle of the Industrial Enzyme alpha-Amino-epsilon-Caprolactam Racemase (ACLR) Observed Using X-ray Crystallography
Acs Catalysis, 7, 2017
|
|
2WIV
 
 | | Cytochrome-P450 XplA heme domain P21 | | Descriptor: | CYTOCHROME P450-LIKE PROTEIN XPLA, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sabbadin, F, Jackson, R, Bruce, N.C, Grogan, G. | | Deposit date: | 2009-05-18 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.5-A Structure of Xpla-Heme, an Unusual Cytochrome P450 Heme Domain that Catalyzes Reductive Biotransformation of Royal Demolition Explosive.
J.Biol.Chem., 284, 2009
|
|
2J5S
 
 | | Structural of ABDH, a beta-diketone hydrolase from the Cyanobacterium Anabaena sp. PCC 7120 bound to (S)-3-oxocyclohexyl acetic acid | | Descriptor: | (S)-CYCLOHEXANONE-2-ACETATE, BETA-DIKETONE HYDROLASE, NICKEL (II) ION | | Authors: | Bennett, J.P, Whittingham, J.L, Brzozowski, A.M, Leonard, P.M, Grogan, G. | | Deposit date: | 2006-09-19 | | Release date: | 2007-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Characterisation of a Beta Diketone Hydrolase from the Cyanobacterium Anabaena Sp. Pcc 7120 in Native and Product Bound Forms, a Coenzyme A-Independent Member of the Crotonase Suprafamily
Biochemistry, 46, 2007
|
|
2J5I
 
 | | Crystal Structure of Hydroxycinnamoyl-CoA Hydratase-Lyase | | Descriptor: | P-HYDROXYCINNAMOYL COA HYDRATASE/LYASE | | Authors: | Leonard, P.M, Brzozowski, A.M, Lebedev, A, Marshall, C.M, Smith, D.J, Verma, C.S, Walton, N.J, Grogan, G. | | Deposit date: | 2006-09-18 | | Release date: | 2006-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A Resolution Structure of Hydroxycinnamoyl- Coenzyme a Hydratase-Lyase (Hchl) from Pseudomonas Fluorescens, an Enzyme that Catalyses the Transformation of Feruloyl-Coenzyme a to Vanillin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2J5G
 
 | | The Native structure of a beta-Diketone Hydrolase from the Cyanobacterium Anabaena sp. PCC 7120 | | Descriptor: | ALR4455 PROTEIN, SULFATE ION | | Authors: | Bennett, J.P, Whittingham, J.L, Brzozowski, A.M, Leonard, P.M, Grogan, G. | | Deposit date: | 2006-09-18 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Characterisation of a Beta Diketone Hydrolase from the Cyanobacterium Anabaena Sp. Pcc 7120 in Native and Product Bound Forms, a Coenzyme A-Independent Member of the Crotonase Suprafamily
Biochemistry, 46, 2007
|
|
6RYZ
 
 | | SalL with S-adenosyl methionine | | Descriptor: | 1,2-ETHANEDIOL, Adenosyl-chloride synthase, CHLORIDE ION, ... | | Authors: | McKean, I, Frese, A, Cuetos, A, Burley, G, Grogan, G. | | Deposit date: | 2019-06-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | S-Adenosyl Methionine Cofactor Modifications Enhance the Biocatalytic Repertoire of Small Molecule C-Alkylation.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6RZ2
 
 | | SalL with Chloroadenosine | | Descriptor: | 5'-CHLORO-5'-DEOXYADENOSINE, Adenosyl-chloride synthase | | Authors: | McKean, I, Frese, A, Cuetos, A, Burley, G, Grogan, G. | | Deposit date: | 2019-06-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | S-Adenosyl Methionine Cofactor Modifications Enhance the Biocatalytic Repertoire of Small Molecule C-Alkylation.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
8P2J
 
 | |
8OZV
 
 | | Imine Reductase from Ajellomyces dermatitidis in complex with 2,2-difluoroacetophenone | | Descriptor: | 2,2-bis(fluoranyl)-1-phenyl-ethanone, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase | | Authors: | Sharma, M, Grogan, G. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure of the imine reductase from Ajellomyces dermatitidis in three crystal forms.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8OZW
 
 | | Imine Reductase from Ajellomyces dermatitidis in complex NADPH4 | | Descriptor: | 1,2-ETHANEDIOL, 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, Oxidoreductase | | Authors: | Sharma, M, Grogan, G. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the imine reductase from Ajellomyces dermatitidis in three crystal forms.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8PPP
 
 | |
8PYY
 
 | |
8PYX
 
 | |
3ZGJ
 
 | |
5A9T
 
 | | Imine Reductase from Amycolatopsis orientalis in complex with (R)- Methyltetrahydroisoquinoline | | Descriptor: | (1R)-1-methyl-1,2,3,4-tetrahydroisoquinoline, ACETATE ION, CALCIUM ION, ... | | Authors: | Man, H, Aleku, G, Turner, N.J, Grogan, G. | | Deposit date: | 2015-07-22 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stereoselectivity and Structural Characterization of an Imine Reductase (Ired) from Amycolatopsis Orientalis
Acs Catalysis, 6, 2016
|
|
5A9R
 
 | | Apo form of Imine reductase from Amycolatopsis orientalis | | Descriptor: | ACETATE ION, IMINE REDUCTASE | | Authors: | Man, H, Aleku, G, Turner, N.J, Grogan, G. | | Deposit date: | 2015-07-22 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Stereoselectivity and Structural Characterization of an Imine Reductase (Ired) from Amycolatopsis Orientalis
Acs Catalysis, 6, 2016
|
|
