2GIC
 
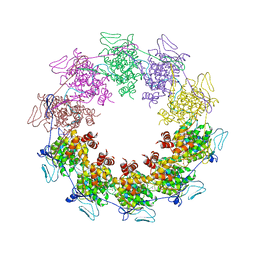 | | Crystal Structure of a vesicular stomatitis virus nucleocapsid-RNA complex | | Descriptor: | 45-MER, Nucleocapsid protein, URANYL (VI) ION | | Authors: | Green, T.J, Zhang, X, Wertz, G.W, Luo, M. | | Deposit date: | 2006-03-28 | | Release date: | 2006-08-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structure of the vesicular stomatitis virus nucleoprotein-RNA complex unveils how the RNA is sequestered
Science, 313, 2006
|
|
7JXR
 
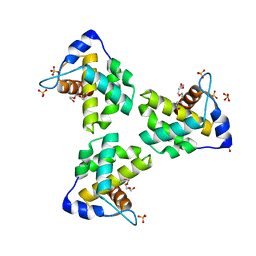 | | Crystal Structure Human Immunodeficiency Virus-1 Matrix protein Mutant Q63R Crystal Form 1 | | Descriptor: | HEXAETHYLENE GLYCOL, Matrix protein, SULFATE ION | | Authors: | Green, T.J, Eastep, G.N, Ghanam, R.H, Saad, J.S. | | Deposit date: | 2020-08-27 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural characterization of HIV-1 matrix mutants implicated in envelope incorporation.
J.Biol.Chem., 296, 2021
|
|
7JXS
 
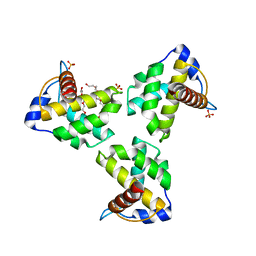 | | Crystal Structure Human Immunodeficiency Virus-1 Matrix protein Mutant Q63R Crystal Form 2 | | Descriptor: | ACETATE ION, DODECAETHYLENE GLYCOL, HEXAETHYLENE GLYCOL, ... | | Authors: | Green, T.J, Eastep, G.N, Ghanam, R.H, Saad, J.S. | | Deposit date: | 2020-08-27 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of HIV-1 matrix mutants implicated in envelope incorporation.
J.Biol.Chem., 296, 2021
|
|
7TBP
 
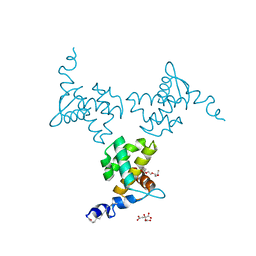 | | X-ray structure of the HIV-1 myristoylated matrix protein | | Descriptor: | CITRIC ACID, Capsid protein p24, MYRISTIC ACID, ... | | Authors: | Green, T.J, Saad, J.S, Samal, A.B. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Atomic view of the HIV-1 matrix lattice; implications on virus assembly and envelope incorporation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7K7P
 
 | |
3HHW
 
 | |
3HHZ
 
 | |
3CRK
 
 | | Crystal structure of the PDHK2-L2 complex. | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial, POTASSIUM ION, ... | | Authors: | Green, T.J, Popov, K.M, Luo, M, Grigorian, A, Klyuyeva, A, Tuganova, A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-04-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional insights into the molecular mechanisms responsible for the regulation of pyruvate dehydrogenase kinase 2.
J.Biol.Chem., 283, 2008
|
|
5CHS
 
 | |
2QVJ
 
 | | Crystal structure of a vesicular stomatitis virus nucleocapsid protein Ser290Trp mutant | | Descriptor: | Nucleocapsid protein | | Authors: | Luo, M, Green, T.J, Zhang, X, Tsao, J, Qiu, S. | | Deposit date: | 2007-08-08 | | Release date: | 2008-01-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Role of intermolecular interactions of vesicular stomatitis virus nucleoprotein in RNA encapsidation.
J.Virol., 82, 2008
|
|
3BXW
 
 | |
2FQM
 
 | |
4O6G
 
 | | Rv3902c from M. tuberculosis | | Descriptor: | Uncharacterized protein | | Authors: | Reddy, B.G, Moates, D.B, Kim, H, Green, T.J, Kim, C, Terwilliger, T.J, Delucas, L.J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-12-20 | | Release date: | 2014-03-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 angstrom resolution X-ray crystal structure of Rv3902c from Mycobacterium tuberculosis.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
3ZLA
 
 | | Crystal structure of the nucleocapsid protein from Bunyamwera virus bound to RNA | | Descriptor: | NUCLEOPROTEIN, RNA | | Authors: | Ariza, A, Tanner, S.J, Walter, C.T, Dent, K.C, Shepherd, D.A, Wu, W, Matthews, S.V, Hiscox, J.A, Green, T.J, Luo, M, Elliot, R.M, Ashcroft, A.E, Stonehouse, N.J, Ranson, N.A, Barr, J.N, Edwards, T.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Nucleocapsid Protein Structures from Orthobunyaviruses Reveal Insight Into Ribonucleoprotein Architecture and RNA Polymerization.
Nucleic Acids Res., 41, 2013
|
|
3ZL9
 
 | | Crystal structure of the nucleocapsid protein from Schmallenberg virus | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Ariza, A, Tanner, S.J, Walter, C.T, Dent, K.C, Shepherd, D.A, Wu, W, Matthews, S.V, Hiscox, J.A, Green, T.J, Luo, M, Elliot, R.M, Ashcroft, A.E, Stonehouse, N.J, Ranson, N.A, Barr, J.N, Edwards, T.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Nucleocapsid Protein Structures from Orthobunyaviruses Reveal Insight Into Ribonucleoprotein Architecture and RNA Polymerization.
Nucleic Acids Res., 41, 2013
|
|
5FTH
 
 | |
5FTI
 
 | |
6DGK
 
 | |
3CRL
 
 | | Crystal structure of the PDHK2-L2 complex. | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial, MAGNESIUM ION, ... | | Authors: | Popov, K.M, Luo, M, Green, T.J, Grigorian, A, Klyuyeva, A, Tuganova, A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-04-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural and functional insights into the molecular mechanisms responsible for the regulation of pyruvate dehydrogenase kinase 2.
J.Biol.Chem., 283, 2008
|
|
1YAE
 
 | | Structure of the Kainate Receptor Subunit GluR6 Agonist Binding Domain Complexed with Domoic Acid | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor, ... | | Authors: | Nanao, M.H, Green, T, Stern-Bach, Y, Heinemann, S.F, Choe, S. | | Deposit date: | 2004-12-17 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure of the kainate receptor subunit GluR6 agonist-binding domain complexed with domoic acid.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2WYY
 
 | | CRYOEM MODEL OF THE VESICULAR STOMATITIS VIRUS | | Descriptor: | NUCLEOPROTEIN, POLY-URIDINE | | Authors: | Ge, P, Tsao, J, Green, T.J, Luo, M, Zhou, Z.H. | | Deposit date: | 2009-11-20 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.6 Å) | | Cite: | Cryo-Em Model of the Bullet-Shaped Vesicular Stomatitis Virus.
Science, 327, 2010
|
|
4EIJ
 
 | |
4BDQ
 
 | | Crystal structure of the GluK2 R775A LBD dimer in complex with glutamate | | Descriptor: | GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, GLUTAMIC ACID, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2012-10-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Correlating Efficacy and Desensitization with Gluk2 Ligand-Binding Domain Movements.
Open Biol., 3, 2013
|
|
2XXT
 
 | | Crystal structure of the GluK2 (GluR6) wild-type LBD dimer in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, CHLORIDE ION, GLUTAMATE RECEPTOR, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational Flexibility of the Ligand-Binding Domain Dimer in Kainate Receptor Gating and Desensitization
J.Neurosci., 31, 2011
|
|
2XXR
 
 | | Crystal structure of the GluK2 (GluR6) wild-type LBD dimer in complex with glutamate | | Descriptor: | CHLORIDE ION, GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2010-11-11 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational Flexibility of the Ligand-Binding Domain Dimer in Kainate Receptor Gating and Desensitization
J.Neurosci., 31, 2011
|
|
