4XP5
 
 | | X-ray structure of Drosophila dopamine transporter bound to cocaine analogue-RTI55 | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Antibody fragment heavy chain-protein, 9D5-heavy chain, ... | | Authors: | Gouaux, E, Penmatsa, A, Wang, K. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
1FTM
 
 | |
4XP1
 
 | | X-ray structure of Drosophila dopamine transporter bound to neurotransmitter dopamine | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gouaux, E, Penmatsa, A, Wang, K. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
1FTK
 
 | |
2NWW
 
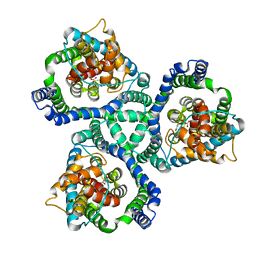 | | Crystal structure of GltPh in complex with TBOA | | Descriptor: | (3S)-3-(BENZYLOXY)-L-ASPARTIC ACID, 425aa long hypothetical proton glutamate symport protein | | Authors: | Gouaux, E, Boudker, O, Ryan, R, Yernool, D, Shimamoto, K. | | Deposit date: | 2006-11-16 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter.
Nature, 445, 2007
|
|
4TLL
 
 | | Crystal structure of GluN1/GluN2B NMDA receptor, structure 1 | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, ... | | Authors: | Gouaux, E, Lee, C.-H, Lu, W. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | NMDA receptor structures reveal subunit arrangement and pore architecture.
Nature, 511, 2014
|
|
4TLM
 
 | | Crystal structure of GluN1/GluN2B NMDA receptor, structure 2 | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, ... | | Authors: | Gouaux, E, Lee, C.-H, Lu, W. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | NMDA receptor structures reveal subunit arrangement and pore architecture.
Nature, 511, 2014
|
|
2NWX
 
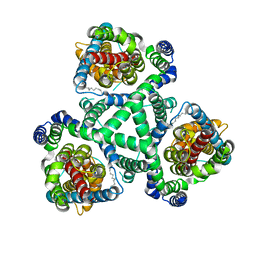 | | Crystal structure of GltPh in complex with L-aspartate and sodium ions | | Descriptor: | 425aa long hypothetical proton glutamate symport protein, ASPARTIC ACID, PALMITIC ACID, ... | | Authors: | Gouaux, E, Boudker, O, Ryan, R, Yernool, D, Shimamoto, K. | | Deposit date: | 2006-11-16 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter.
Nature, 445, 2007
|
|
2NWL
 
 | | Crystal structure of GltPh in complex with L-Asp | | Descriptor: | ASPARTIC ACID, PALMITIC ACID, glutamate symport protein | | Authors: | Gouaux, E, Boudker, O, Ryan, R, Yernool, D, Shimamoto, K. | | Deposit date: | 2006-11-15 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter.
Nature, 445, 2007
|
|
6C13
 
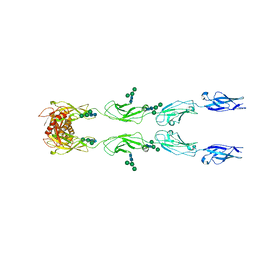 | | CryoEM structure of mouse PCDH15-4EC-LHFPL5 complex | | Descriptor: | Protocadherin-15, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gouaux, E, Ge, J, Elferich, J. | | Deposit date: | 2018-01-03 | | Release date: | 2018-08-15 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (11.33 Å) | | Cite: | Structure of mouse protocadherin 15 of the stereocilia tip link in complex with LHFPL5.
Elife, 7, 2018
|
|
6C10
 
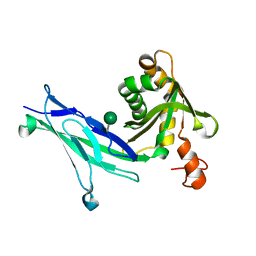 | | Crystal structure of mouse PCDH15 EC11-EL | | Descriptor: | Protocadherin-15, alpha-D-mannopyranose | | Authors: | Gouaux, E, Elferich, J, Ge, J. | | Deposit date: | 2018-01-03 | | Release date: | 2018-08-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structure of mouse protocadherin 15 of the stereocilia tip link in complex with LHFPL5.
Elife, 7, 2018
|
|
6C14
 
 | |
6NJL
 
 | |
6NJN
 
 | |
6NJM
 
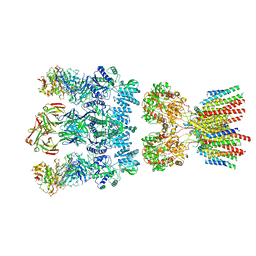 | | Architecture and subunit arrangement of native AMPA receptors | | Descriptor: | 15F1 Fab heavy chain, 15F1 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gouaux, E, Zhao, Y. | | Deposit date: | 2019-01-03 | | Release date: | 2019-04-24 | | Last modified: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM.
Science, 364, 2019
|
|
4M48
 
 | | X-ray structure of dopamine transporter elucidates antidepressant mechanism | | Descriptor: | 9D5 antibody, heavy chain, light chain, ... | | Authors: | Gouaux, E, Penmatsa, A, Wang, K. | | Deposit date: | 2013-08-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | X-ray structure of dopamine transporter elucidates antidepressant mechanism.
Nature, 503, 2013
|
|
2QEI
 
 | | Crystal structure analysis of LeuT complexed with L-alanine, sodium, and clomipramine | | Descriptor: | 3-(3-CHLORO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N,N-DIMETHYLPROPAN-1-AMINE, ALANINE, SODIUM ION, ... | | Authors: | Singh, S.K, Yamashita, A, Gouaux, E. | | Deposit date: | 2007-06-25 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Antidepressant binding site in a bacterial homologue of neurotransmitter transporters.
Nature, 448, 2007
|
|
3TT1
 
 | |
3USG
 
 | | Crystal structure of LeuT bound to L-leucine in space group C2 from lipid bicelles | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, LEUCINE, ... | | Authors: | Wang, H, Elferich, J, Gouaux, E. | | Deposit date: | 2011-11-23 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1N0T
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with the antagonist (S)-ATPO at 2.1 A resolution. | | Descriptor: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Greenwood, J.R, Liljefors, T, Lunn, M.-L, Egebjerg, J, Larsen, I.K, Gouaux, E, Kastrup, J.S. | | Deposit date: | 2002-10-15 | | Release date: | 2003-03-04 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Competitive antagonism of AMPA receptors by ligands of
different classes: crystal structure of ATPO bound to the
GluR2 ligand-binding core, in comparison with DNQX.
J.Med.Chem., 46, 2003
|
|
4MMD
 
 | |
1MS7
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-Des-Me-AMPA at 1.97 A resolution, Crystallization in the presence of zinc acetate | | Descriptor: | (S)-2-AMINO-3-(3-HYDROXY-ISOXAZOL-4-YL)PROPIONIC ACID, Glutamate receptor subunit 2, ZINC ION | | Authors: | Kasper, C, Lunn, M.-L, Liljefors, T, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2002-09-19 | | Release date: | 2003-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | GluR2 ligand-binding core complexes: importance of the isoxazolol moiety and 5-substituent for the binding mode of AMPA-type agonists
FEBS Lett., 531, 2002
|
|
8TKP
 
 | | Structure of the C. elegans TMC-2 complex | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Clark, S, Jeong, H, Goehring, A, Posert, R, Gouaux, E. | | Deposit date: | 2023-07-25 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The structure of the Caenorhabditis elegans TMC-2 complex suggests roles of lipid-mediated subunit contacts in mechanosensory transduction.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4NTW
 
 | | Structure of acid-sensing ion channel in complex with snake toxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, Basic phospholipase A2 homolog Tx-beta, ... | | Authors: | Baconguis, I, Bohlen, C.J, Goehring, A, Julius, D, Gouaux, E. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | X-ray structure of Acid-sensing ion channel 1-snake toxin complex reveals open state of a na(+)-selective channel.
Cell(Cambridge,Mass.), 156, 2014
|
|
1M5E
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH ACPA AT 1.46 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
