1DP8
 
 | |
4Y07
 
 | | Crystal structure of the HECT domain of human WWP2 | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP2 | | Authors: | Gong, W, Li, J, Li, Z, Xu, Y. | | Deposit date: | 2015-02-05 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Structure of the HECT domain of human WWP2
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1DP9
 
 | | CRYSTAL STRUCTURE OF IMIDAZOLE-BOUND FIXL HEME DOMAIN | | Descriptor: | FIXL PROTEIN, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gong, W, Hao, B, Chan, M.K. | | Deposit date: | 1999-12-24 | | Release date: | 2000-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL.
Biochemistry, 39, 2000
|
|
1DP6
 
 | | OXYGEN-BINDING COMPLEX OF FIXL HEME DOMAIN | | Descriptor: | FIXL PROTEIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gong, W, Hao, B, Chan, M.K. | | Deposit date: | 1999-12-23 | | Release date: | 2000-12-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL.
Biochemistry, 39, 2000
|
|
3CF4
 
 | | Structure of the CODH component of the M. barkeri ACDS complex | | Descriptor: | ACETIC ACID, Acetyl-CoA decarboxylase/synthase alpha subunit, Acetyl-CoA decarboxylase/synthase epsilon subunit, ... | | Authors: | Gong, W, Hao, B, Wei, Z, Ferguson Jr, D.J, Tallant, T, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2008-03-01 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the alpha2 epsilon2 Ni-dependent CO dehydrogenase component of the Methanosarcina barkeri acetyl-CoA decarboxylase/synthase complex
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2OZX
 
 | |
2OZW
 
 | |
7EY1
 
 | | Bifunctional xylosidase/glucosidase LXYL with intermediate substrate xylose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(4-3)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-xylosidase/beta-D-glucosidase, ... | | Authors: | Gong, W.M, Yang, L.Y. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Bifunctional xylosidase/glucosidase LXYL with intermediate substrate xylose
To Be Published
|
|
7EY2
 
 | |
1DRM
 
 | | CRYSTAL STRUCTURE OF THE LIGAND FREE BJFIXL HEME DOMAIN | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SENSOR PROTEIN FIXL | | Authors: | Gong, W, Hao, B, Mansy, S.S, Gonzalez, G, Gilles-Gonzalez, M.A, Chan, M.K. | | Deposit date: | 2000-01-06 | | Release date: | 2000-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
8IS0
 
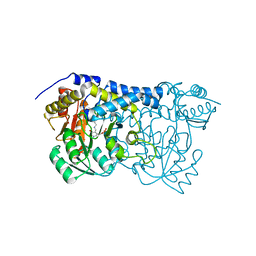 | | Carbon Sulfoxide lyase - Y106F | | Descriptor: | 2-AMINO-ACRYLIC ACID, PYRIDOXAL-5'-PHOSPHATE, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
8IRZ
 
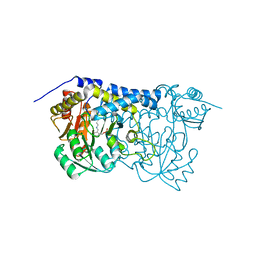 | | Carbon Sulfoxide lyase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
8IRY
 
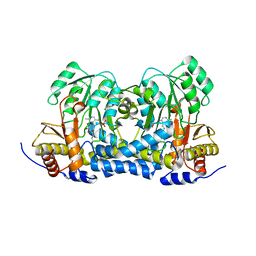 | | Carbon Sulfoxide lyase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, PYRUVIC ACID, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
2AI6
 
 | |
8H7V
 
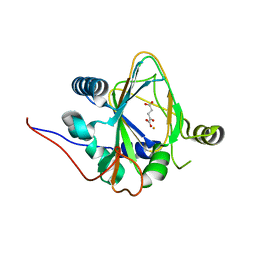 | | Trans-3/4-proline-hydroxylase H11 with AKG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H81
 
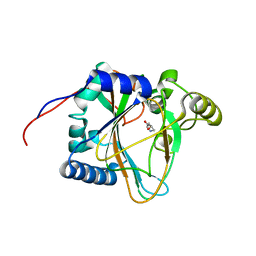 | | Trans-3/4-proline-hydroxylase H11 with 4-Hydroxyl-proline | | Descriptor: | 4-HYDROXYPROLINE, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H85
 
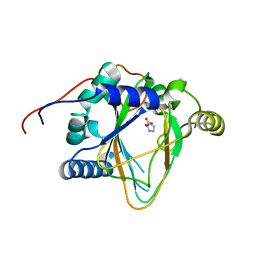 | | Trans-3/4-proline-hydroxylase H11 with 3-hydroxyl-proline | | Descriptor: | 3-HYDROXYPROLINE, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H7Y
 
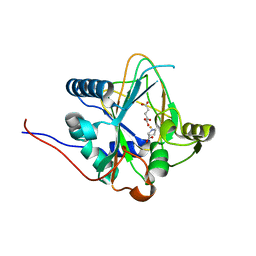 | | Trans-3/4-proline-hydroxylase H11 with AKG and L-proline | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, PROLINE, ... | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H7T
 
 | | Trans-3/4-proline-hydroxylase H11 apo structure | | Descriptor: | CHLORIDE ION, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8WRB
 
 | | Lysophosphatidylserine receptor GPR34-Gi complex | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Gong, W, Liu, G, Li, X, Wang, Y, Zhang, X. | | Deposit date: | 2023-10-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for ligand recognition and signaling of the lysophosphatidylserine receptors GPR34 and GPR174.
Plos Biol., 21, 2023
|
|
1BOO
 
 | | PVUII DNA METHYLTRANSFERASE (CYTOSINE-N4-SPECIFIC) | | Descriptor: | PROTEIN (N-4 CYTOSINE-SPECIFIC METHYLTRANSFERASE PVU II), S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Gong, W, O'Gara, M, Blumenthal, R.M, Cheng, X. | | Deposit date: | 1998-07-31 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of pvu II DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment.
Nucleic Acids Res., 25, 1997
|
|
1BUD
 
 | | ACUTOLYSIN A FROM SNAKE VENOM OF AGKISTRODON ACUTUS AT PH 5.0 | | Descriptor: | CALCIUM ION, PROTEIN (ACUTOLYSIN A), ZINC ION | | Authors: | Gong, W, Zhu, X, Liu, S, Teng, M, Niu, L. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of acutolysin A, a three-disulfide hemorrhagic zinc metalloproteinase from the snake venom of Agkistrodon acutus.
J.Mol.Biol., 283, 1998
|
|
1BSW
 
 | | ACUTOLYSIN A FROM SNAKE VENOM OF AGKISTRODON ACUTUS AT PH 7.5 | | Descriptor: | CALCIUM ION, PROTEIN (ACUTOLYSIN A), ZINC ION | | Authors: | Gong, W, Zhu, X, Liu, S, Teng, M, Niu, L. | | Deposit date: | 1998-08-31 | | Release date: | 1999-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of acutolysin A, a three-disulfide hemorrhagic zinc metalloproteinase from the snake venom of Agkistrodon acutus.
J.Mol.Biol., 283, 1998
|
|
8IRK
 
 | | Carbon Sulfoxide lyase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, PYRUVIC ACID, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-18 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Mycobacterial ergothioneine-biosynthesis C-S lyase EgtE
To Be Published
|
|
8IZB
 
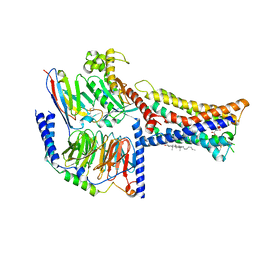 | | Lysophosphatidylserine receptor GPR174-Gs complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gong, W, Liu, G, Li, X, Zhang, X. | | Deposit date: | 2023-04-06 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for ligand recognition and signaling of the lysophosphatidylserine receptors GPR34 and GPR174.
Plos Biol., 21, 2023
|
|
