1L6M
 
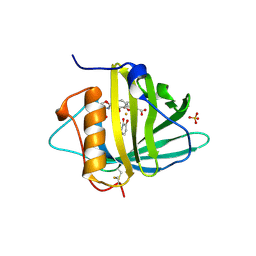 | | Neutrophil Gelatinase-associated Lipocalin is a Novel Bacteriostatic Agent that Interferes with Siderophore-mediated Iron Acquisition | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2-(2,3-DIHYDROXY-BENZOYLAMINO)-3-HYDROXY-PROPIONIC ACID, FE (III) ION, ... | | Authors: | Goetz, D.H, Borregaard, N, Bluhm, M.E, Raymond, K.N, Strong, R.K. | | Deposit date: | 2002-03-11 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Neutrophil Lipocalin NGAL is a Bacteriostatic Agent that Interferes with Siderophore-mediated Iron Acquisition
Mol.Cell, 10, 2002
|
|
1DFV
 
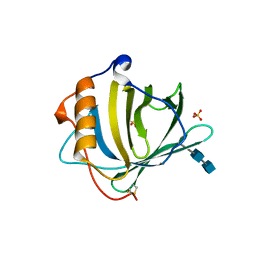 | | CRYSTAL STRUCTURE OF HUMAN NEUTROPHIL GELATINASE ASSOCIATED LIPOCALIN MONOMER | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HUMAN NEUTROPHIL GELATINASE, ... | | Authors: | Goetz, D.H, Willie, S.T, Armen, R.S, Bratt, T, Borregaard, N, Strong, R.K. | | Deposit date: | 1999-11-22 | | Release date: | 2000-03-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ligand preference inferred from the structure of neutrophil gelatinase associated lipocalin
Biochemistry, 39, 2000
|
|
1QQS
 
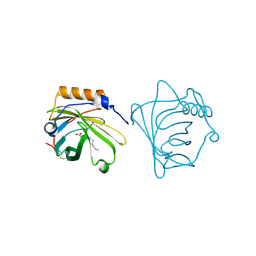 | | NEUTROPHIL GELATINASE ASSOCIATED LIPOCALIN HOMODIMER | | Descriptor: | DECANOIC ACID, NEUTROPHIL GELATINASE, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Goetz, D.H, Willie, S.T, Armen, R, Bratt, T, Borregaard, N, Strong, R.K. | | Deposit date: | 1999-06-07 | | Release date: | 2000-04-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ligand preference inferred from the structure of neutrophil gelatinase associated lipocalin
Biochemistry, 39, 2000
|
|
1ORF
 
 | | The Oligomeric Structure of Human Granzyme A Reveals the Molecular Determinants of Substrate Specificity | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Granzyme A, SULFATE ION | | Authors: | Bell, J.K, Goetz, D.H, Mahrus, S, Harris, J.L, Fletterick, R.J, Craik, C.S. | | Deposit date: | 2003-03-12 | | Release date: | 2003-07-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The oligomeric structure of human granzyme A is a determinant of its extended substrate specificity.
Nat.Struct.Biol., 10, 2003
|
|
3SO3
 
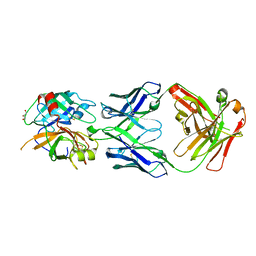 | | Structures of Fab-Protease Complexes Reveal a Highly Specific Non-Canonical Mechanism of Inhibition. | | Descriptor: | A11 FAB heavy chain, A11 FAB light chain, GLYCEROL, ... | | Authors: | Schneider, E.L, Farady, C.J, Egea, P.F, Goetz, D.H, Baharuddin, A, Craik, C.S. | | Deposit date: | 2011-06-29 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A reverse binding motif that contributes to specific protease inhibition by antibodies.
J.Mol.Biol., 415, 2012
|
|
2OP9
 
 | |
2PBK
 
 | |
3BN9
 
 | | Crystal Structure of MT-SP1 in complex with Fab Inhibitor E2 | | Descriptor: | 1,2-ETHANEDIOL, E2 Fab Heavy Chain, E2 Fab Light Chain, ... | | Authors: | Farady, C.J, Schneider, E.L, Egea, P.F, Goetz, D.H, Craik, C.S. | | Deposit date: | 2007-12-13 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.173 Å) | | Cite: | Structure of an Fab-protease complex reveals a highly specific non-canonical mechanism of inhibition
J.Mol.Biol., 380, 2008
|
|
