2H8L
 
 | |
2I3E
 
 | |
1MK3
 
 | | SOLUTION STRUCTURE OF HUMAN BCL-W PROTEIN | | Descriptor: | Apoptosis regulator Bcl-W | | Authors: | Denisov, A.Y, Madiraju, M.S, Chen, G, Khadir, A, Beauparlant, P, Attardo, G, Shore, G.C, Gehring, K. | | Deposit date: | 2002-08-28 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human BCL-w: modulation of ligand binding by the C-terminal helix
J.BIOL.CHEM., 278, 2003
|
|
1NEE
 
 | | Structure of archaeal translation factor aIF2beta from Methanobacterium thermoautrophicum | | Descriptor: | Probable translation initiation factor 2 beta subunit, ZINC ION | | Authors: | Gutierrez, P, Trempe, J.F, Siddiqui, N, Arrowsmith, C, Gehring, K. | | Deposit date: | 2002-12-11 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the archaeal translation initiation factor aIF2beta from Methanobacterium thermoautotrophicum: Implications for translation initiation.
Protein Sci., 13, 2004
|
|
1NMR
 
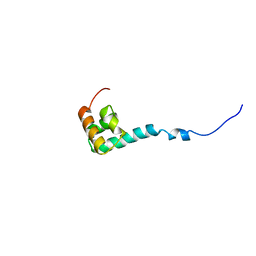 | | Solution Structure of C-terminal Domain from Trypanosoma cruzi Poly(A)-Binding Protein | | Descriptor: | poly(A)-binding protein | | Authors: | Siddiqui, N, Kozlov, G, D'Orso, I, Trempe, J.F, Frasch, A.C.C, Gehring, K. | | Deposit date: | 2003-01-10 | | Release date: | 2003-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain from poly(A)-binding protein in Trypanosoma cruzi: A vegetal PABC domain
Protein Sci., 12, 2003
|
|
1P9K
 
 | | THE SOLUTION STRUCTURE OF YBCJ FROM E. COLI REVEALS A RECENTLY DISCOVERED ALFAL MOTIF INVOLVED IN RNA-BINDING | | Descriptor: | orf, hypothetical protein | | Authors: | Volpon, L, Lievre, C, Osborne, M.J, Gandhi, S, Iannuzzi, P, Larocque, R, Matte, A, Cygler, M, Gehring, K, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-05-12 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of YbcJ from Escherichia coli reveals a recently discovered alphaL motif involved in RNA binding.
J.Bacteriol., 185, 2003
|
|
3IQL
 
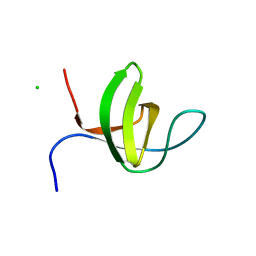 | | Crystal structure of the rat endophilin-A1 SH3 domain | | Descriptor: | CHLORIDE ION, Endophilin-A1 | | Authors: | Trempe, J.F, Kozlov, G, Camacho, E.M, Gehring, K. | | Deposit date: | 2009-08-20 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | SH3 domains from a subset of BAR proteins define a Ubl-binding domain and implicate parkin in synaptic ubiquitination.
Mol.Cell, 36, 2009
|
|
3UVT
 
 | |
7M1U
 
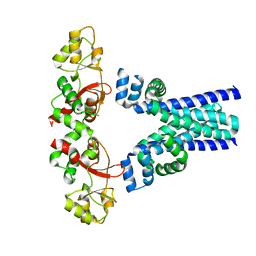 | | Crystal structure of an archaeal CNNM, MtCorB, R235L mutant with C-terminal deletion | | Descriptor: | Hemolysin, contains CBS domains | | Authors: | Chen, Y.S, Kozlov, G, Gehring, K. | | Deposit date: | 2021-03-15 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of an archaeal CorB magnesium transporter.
Nat Commun, 12, 2021
|
|
7MSU
 
 | | Crystal structure of an archaeal CNNM, MtCorB, CBS-pair domain in complex with Mg2+-ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Hemolysin, ... | | Authors: | Chen, Y.S, Gehring, K. | | Deposit date: | 2021-05-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of an archaeal CorB magnesium transporter.
Nat Commun, 12, 2021
|
|
3PDZ
 
 | |
3RG0
 
 | | Structural and functional relationships between the lectin and arm domains of calreticulin | | Descriptor: | CALCIUM ION, Calreticulin | | Authors: | Kozlov, G, Pocanschi, C.L, Brockmeier, U, Williams, D.B, Gehring, K. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural and Functional Relationships between the Lectin and Arm Domains of Calreticulin.
J.Biol.Chem., 286, 2011
|
|
1L1P
 
 | | Solution Structure of the PPIase Domain from E. coli Trigger Factor | | Descriptor: | trigger factor | | Authors: | Kozlov, G, Trempe, J.-F, Perreault, A, Wong, M, Denisov, A, Ghandi, S, Gehring, K, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-02-19 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Closed Form of a Peptidyl-Prolyl Isomerase Reveals the Mechanism of Protein Folding
To be Published
|
|
2OOB
 
 | |
3PT3
 
 | | Crystal structure of the C-terminal lobe of the human UBR5 HECT domain | | Descriptor: | E3 ubiquitin-protein ligase UBR5 | | Authors: | Matta-Camacho, E, Kozlov, G, Menade, M, Gehring, K. | | Deposit date: | 2010-12-02 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of the HECT C-lobe of the UBR5 E3 ubiquitin ligase.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4EF0
 
 | |
4F26
 
 | |
4F25
 
 | |
4GWR
 
 | |
1DU6
 
 | |
1JGN
 
 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip2 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein 2 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-26 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
1JH4
 
 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip1 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein-1 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-27 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
4I6X
 
 | |
2ILX
 
 | |
2K18
 
 | | Solution structure of bb' domains of human protein disulfide isomerase | | Descriptor: | Protein disulfide-isomerase | | Authors: | Denisov, A.Y, Maattanen, P, Dabrowski, C, Kozlov, G, Thomas, D.Y, Gehring, K. | | Deposit date: | 2008-02-22 | | Release date: | 2008-04-29 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the bb' domains of human protein disulfide isomerase.
Febs J., 276, 2009
|
|
