5IIN
 
 | | Crystal structure of the pre-catalytic ternary extension complex of DNA polymerase lambda with an 8-oxo-dG:dC base-pair | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*(8OG)P*TP*AP*CP*TP*G)-3'), ... | | Authors: | Burak, M.J, Guja, K.E, Garcia-Diaz, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A fidelity mechanism in DNA polymerase lambda promotes error-free bypass of 8-oxo-dG.
Embo J., 35, 2016
|
|
5IIK
 
 | | Crystal structure of the post-catalytic nick complex of DNA polymerase lambda with a templating 8-oxo-dG and incorporated dC | | Descriptor: | CITRIC ACID, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*(8OG)P*TP*AP*CP*TP*G)-3'), ... | | Authors: | Burak, M.J, Guja, K.E, Garcia-Diaz, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | A fidelity mechanism in DNA polymerase lambda promotes error-free bypass of 8-oxo-dG.
Embo J., 35, 2016
|
|
5III
 
 | | Crystal structure of the pre-catalytic ternary complex of DNA polymerase lambda with a templating 8-oxo-dG and an incoming dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*AP*(DOC))-3'), DNA (5'-D(*CP*GP*GP*CP*(8OG)P*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Burak, M.J, Guja, K.E, Garcia-Diaz, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A fidelity mechanism in DNA polymerase lambda promotes error-free bypass of 8-oxo-dG.
Embo J., 35, 2016
|
|
4OHU
 
 | | Crystal structure of Mycobacterium tuberculosis InhA in complex with inhibitor PT92 | | Descriptor: | 2-(2-bromophenoxy)-5-hexylphenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Yu, W, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-01-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
5CKY
 
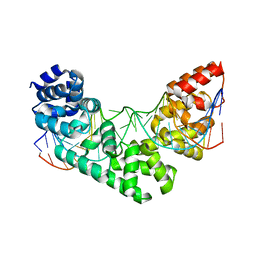 | | Crystal Structure of the MTERF1 R162A substitution bound to the termination sequence. | | Descriptor: | 5' -D (*TP*AP*AP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*GP*TP*AP*AP*T)-3', 5'-D(*AP*TP*TP*AP*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*CP*AP*TP*CP*TP*TP*A)-3', Transcription termination factor 1, ... | | Authors: | Byrnes, J, Hauser, K, Norona, L, Mejia, E, Simmerling, C, Garcia-Diaz, M. | | Deposit date: | 2015-07-15 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Base Flipping by MTERF1 Can Accommodate Multiple Conformations and Occurs in a Stepwise Fashion.
J.Mol.Biol., 428, 2016
|
|
5CRK
 
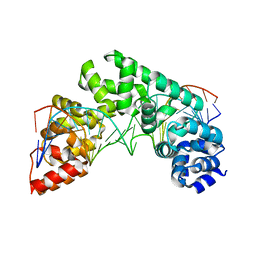 | | Crystal Structure of the MTERF1 F243A substitution bound to the termination sequence. | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*CP*AP*TP*CP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*GP*TP*AP*AP*T)-3'), Transcription termination factor 1, ... | | Authors: | Byrnes, J, Hauser, K, Norona, L, Mejia, E, Simmerling, C, Garcia-Diaz, M. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Base Flipping by MTERF1 Can Accommodate Multiple Conformations and Occurs in a Stepwise Fashion.
J.Mol.Biol., 428, 2016
|
|
5CRJ
 
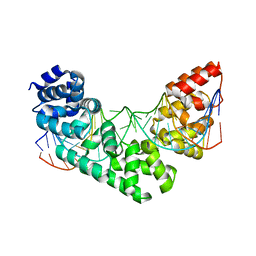 | | Crystal Structure of the MTERF1 F322A substitution bound to the termination sequence. | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*CP*AP*TP*CP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*GP*TP*AP*AP*T)-3'), Transcription termination factor 1, ... | | Authors: | Byrnes, J, Hauser, K, Norona, L, Mejia, E, Simmerling, C, Garcia-Diaz, M. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Base Flipping by MTERF1 Can Accommodate Multiple Conformations and Occurs in a Stepwise Fashion.
J.Mol.Biol., 428, 2016
|
|
3LO5
 
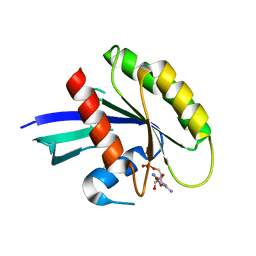 | | Crystal Structure of the dominant negative S17N mutant of Ras | | Descriptor: | CALCIUM ION, GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nassar, N, Singh, K, Garcia-Diaz, M. | | Deposit date: | 2010-02-03 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | Structure of the Dominant Negative S17N Mutant of Ras
Biochemistry, 49, 2010
|
|
3T8B
 
 | | Crystal structure of Mycobacterium tuberculosis MenB with altered hexameric assembly | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, GLYCEROL | | Authors: | Li, H.-J, Li, X, Liu, N, Zhang, H, Truglio, J, Mishra, S, Kisker, C, Garcia-Diaz, M, Tonge, P. | | Deposit date: | 2011-08-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Mechanism of the Intramolecular Claisen Condensation Reaction Catalyzed by MenB, a Crotonase Superfamily Member.
Biochemistry, 50, 2011
|
|
3T8A
 
 | | Crystal structure of Mycobacterium tuberculosis MenB in complex with substrate analogue, OSB-NCoA | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, o-succinylbenzoyl-N-coenzyme A | | Authors: | Li, H.-J, Li, X, Liu, N, Zhang, H, Truglio, J, Mishra, S, Kisker, C, Garcia-Diaz, M, Tonge, P. | | Deposit date: | 2011-08-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Mechanism of the Intramolecular Claisen Condensation Reaction Catalyzed by MenB, a Crotonase Superfamily Member.
Biochemistry, 50, 2011
|
|
4P4P
 
 | | Crystal structure of Leishmania infantum polymerase beta: Nick complex | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(P*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), DNA (5'-D(P*GP*CP*CP*G)-3'), ... | | Authors: | Mejia, E, Burak, M, Alonso, A, Larraga, V, Kunkel, T, Bebenek, K, Garcia-Diaz, M. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2973 Å) | | Cite: | Structures of the Leishmania infantum polymerase beta.
DNA Repair (Amst.), 18, 2014
|
|
5BMM
 
 | | Src in complex with DNA-templated macrocyclic inhibitor MC25b | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, macrocyclic inhibitor MC25b | | Authors: | Georghiou, G, Guja, K.E, Aleem, S, Kleiner, R.E, Liu, D.R, Miller, W.T, Garcia-Diaz, M, Seeliger, M.A. | | Deposit date: | 2015-05-22 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Basis for Intracellular Kinase Inhibition by Src-specific Peptidic Macrocycles.
Cell Chem Biol, 23, 2016
|
|
4P4O
 
 | | Crystal structure of Leishmania infantum polymerase beta: Ternary gap complex | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*AP*T)-3'), DNA (5'-D(P*CP*GP*GP*CP*AP*AP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Mejia, E, Burak, M, Alonso, A, Larraga, V, Kunkel, T, Bebenek, K, Garcia-Diaz, M. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3001 Å) | | Cite: | Structures of the Leishmania infantum polymerase beta.
DNA Repair (Amst.), 18, 2014
|
|
4P4M
 
 | | Crystal structure of Leishmania infantum polymerase beta: Ternary P/T complex | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*A)-3'), DNA (5'-D(P*AP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Mejia, E, Burak, M, Alonso, A, Larraga, V, Kunkel, T.A, Bebenek, K, Garcia-Diaz, M. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9185 Å) | | Cite: | Structures of the Leishmania infantum polymerase beta.
DNA Repair (Amst.), 18, 2014
|
|
5COQ
 
 | | The effect of valine to alanine mutation on InhA enzyme crystallization pattern and substrate binding loop conformation and flexibility | | Descriptor: | 5-HEXYL-2-(2-METHYLPHENOXY)PHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Li, H.-J, Lai, C.-T, Liu, N, Yu, W, Shah, S, Bommineni, G.R, Perrone, V, Garcia-Diaz, M, Tonge, P.J, Simmerling, C. | | Deposit date: | 2015-07-20 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Modulation of the Induced-Fit Conformational Change for Slow-Onset Inhibition in Mycobacterium tuberculosis InhA.
Biochemistry, 54, 2015
|
|
5CPB
 
 | | The effect of isoleucine to alanine mutation on InhA enzyme crystallization pattern and inhibition by ligand PT70 (TCU) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.-J, Lai, C.-T, Liu, N, Yu, W, Shah, S, Bommineni, G.R, Perrone, V, Garcia-Diaz, M, Tonge, P.J, Simmerling, C. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Rational Modulation of the Induced-Fit Conformational Change for Slow-Onset Inhibition in Mycobacterium tuberculosis InhA.
Biochemistry, 54, 2015
|
|
4OXN
 
 | | Substrate-like binding mode of inhibitor PT155 to the Mycobacterium tuberculosis enoyl-ACP reductase InhA | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-(4-amino-2-methylphenoxy)-2-hexyl-4-hydroxy-1-methylpyridinium, ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-05 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2926 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OXK
 
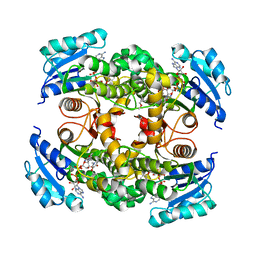 | | Multiple binding modes of inhibitor PT155 to the Mycobacterium tuberculosis enoyl-ACP reductase InhA within a tetramer | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, 5-(4-amino-2-methylphenoxy)-2-hexyl-4-hydroxy-1-methylpyridinium, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-05 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8429 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OYR
 
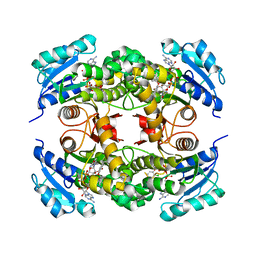 | | Competition of the small inhibitor PT91 with large fatty acyl substrate of the Mycobacterium tuberculosis enoyl-ACP reductase InhA by induced substrate-binding loop refolding | | Descriptor: | 2-(2-chloranylphenoxy)-5-hexyl-phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2995 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OXY
 
 | | Substrate-binding loop movement with inhibitor PT10 in the tetrameric Mycobacterium tuberculosis enoyl-ACP reductase InhA | | Descriptor: | 5-hexyl-2-(2-nitrophenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Sullivan, T.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-09 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
3MVB
 
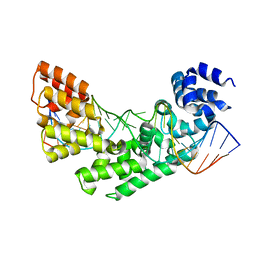 | | Crystal structure of a triple RFY mutant of human MTERF1 bound to the termination sequence | | Descriptor: | 5'-D(*AP*TP*TP*AP*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*CP*AP*TP*CP*TP*TP*A)-3', 5'-D(*TP*AP*AP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*GP*TP*AP*AP*T)-3', Transcription termination factor, ... | | Authors: | Yakubovskaya, E, Mejia, E, Byrnes, J, Hambardjieva, E, Garcia-Diaz, M. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.786 Å) | | Cite: | Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription.
Cell(Cambridge,Mass.), 141, 2010
|
|
3MVA
 
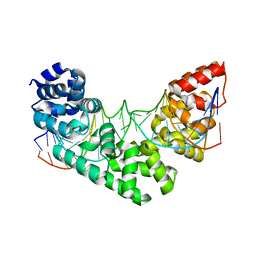 | | Crystal structure of human MTERF1 bound to the termination sequence | | Descriptor: | 5'-D(*AP*TP*TP*AP*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*CP*AP*TP*CP*TP*TP*A)-3'), 5'-D(*TP*AP*AP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*GP*TP*AP*AP*T)-3'), Transcription termination factor, ... | | Authors: | Yakubovskaya, E, Mejia, E, Byrnes, J, Hambardjieva, E, Garcia-Diaz, M. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription.
Cell(Cambridge,Mass.), 141, 2010
|
|
5CPF
 
 | | Compensation of the effect of isoleucine to alanine mutation by designed inhibition in the InhA enzyme | | Descriptor: | 2-(2-methylphenoxy)-5-[(4-phenyl-1H-1,2,3-triazol-1-yl)methyl]phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.-J, Lai, C.-T, Pan, P, Yu, W, Shah, S, Bommineni, G.R, Perrone, V, Garcia-Diaz, M, Tonge, P.J, Simmerling, C. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.409 Å) | | Cite: | Rational Modulation of the Induced-Fit Conformational Change for Slow-Onset Inhibition in Mycobacterium tuberculosis InhA.
Biochemistry, 54, 2015
|
|
5CP8
 
 | | The effect of isoleucine to alanine mutation on InhA enzyme crystallization pattern and substrate binding loop conformation and flexibility | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-HEXYL-2-(2-METHYLPHENOXY)PHENOL, ... | | Authors: | Li, H.-J, Lai, C.-T, Liu, N, Yu, W, Shah, S, Bommineni, G.R, Perrone, V, Garcia-Diaz, M, Tonge, P.J, Simmerling, C. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational Modulation of the Induced-Fit Conformational Change for Slow-Onset Inhibition in Mycobacterium tuberculosis InhA.
Biochemistry, 54, 2015
|
|
