2R3H
 
 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 3-methyl-N-(pyridin-4-ylmethyl)imidazo[1,2-a]pyrazin-8-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3J
 
 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 3-bromo-5-phenyl-N-(pyridin-3-ylmethyl)pyrazolo[1,5-a]pyrimidin-7-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3L
 
 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 3-bromo-6-phenyl-N-(pyrimidin-5-ylmethyl)imidazo[1,2-a]pyridin-8-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3I
 
 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 5-(2-fluorophenyl)-N-(pyridin-4-ylmethyl)pyrazolo[1,5-a]pyrimidin-7-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3O
 
 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | (5-phenyl-7-(pyridin-3-ylmethylamino)pyrazolo[1,5-a]pyrimidin-3-yl)methanol, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3F
 
 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 5-(2,3-dichlorophenyl)-N-(pyridin-4-ylmethyl)pyrazolo[1,5-a]pyrimidin-7-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3M
 
 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | Cell division protein kinase 2, N-((2-aminopyrimidin-5-yl)methyl)-5-(2,6-difluorophenyl)-3-ethylpyrazolo[1,5-a]pyrimidin-7-amine | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
3PA3
 
 | | X-ray crystal structure of compound 70 bound to human CHK1 kinase domain | | Descriptor: | 2-(4-chlorophenyl)-4-[(3S)-piperidin-3-ylamino]thieno[2,3-d]pyridazine-7-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design, synthesis and SAR of thienopyridines as potent CHK1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3PA4
 
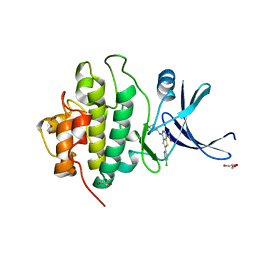 | | X-ray crystal structure of compound 2a bound to human CHK1 kinase domain | | Descriptor: | 2-(4-chlorophenyl)-4-[(3S)-piperidin-3-ylamino]thieno[3,2-c]pyridine-7-carboxamide, GLYCEROL, Serine/threonine-protein kinase Chk1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Design, synthesis and SAR of thienopyridines as potent CHK1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3PA5
 
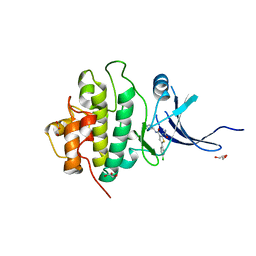 | | X-ray crystal structure of compound 1 bound to human CHK1 kinase domain | | Descriptor: | 2-(carbamoylamino)-5-(4-chlorophenyl)-N-[(3S)-piperidin-3-yl]thiophene-3-carboxamide, GLYCEROL, Serine/threonine-protein kinase Chk1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, synthesis and SAR of thienopyridines as potent CHK1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6BFB
 
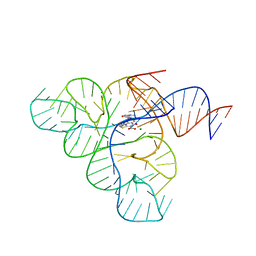 | | Crystal structure of a F. nucleatum FMN riboswitch bound to WG-3 | | Descriptor: | 5-[(3S,4S)-3-(dimethylamino)-4-hydroxypyrrolidin-1-yl]-6-fluoro-4-methyl-8-oxo-3,4-dihydro-8H-1-thia-4,9b-diazacyclopenta[cd]phenalene-9-carboxylic acid, MAGNESIUM ION, RNA (54-MER), ... | | Authors: | Rizvi, N.F, Fischmann, T.O. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery of Selective RNA-Binding Small Molecules by Affinity-Selection Mass Spectrometry.
ACS Chem. Biol., 13, 2018
|
|
7T4W
 
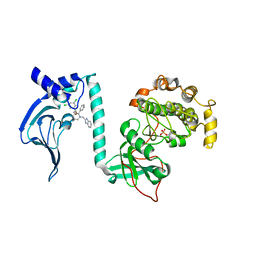 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | (2R)-2-({(2S,3S)-1-[(1H-benzimidazol-2-yl)methyl]-2-phenylpiperidin-3-yl}oxy)-2-(3,5-dichlorophenyl)ethan-1-ol, CHLORIDE ION, cGMP-dependent protein kinase 1 | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
7T4V
 
 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | 4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)-3-methylbenzoic acid, CHLORIDE ION, cGMP-dependent protein kinase 1 | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
7T4U
 
 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | 3-amino-4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)benzoic acid, cGMP-dependent protein kinase 1 | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
7T4T
 
 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
1TLF
 
 | |
4MBI
 
 | | Discovery of Pyrazolo[1,5a]pyrimidine-based Pim1 Inhibitors | | Descriptor: | N,N-dimethyl-N'-[3-(1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrimidin-5-yl]ethane-1,2-diamine, Serine/threonine-protein kinase pim-1 | | Authors: | Azevedo, R, Fischmann, T.O. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of pyrazolo[1,5-a]pyrimidine-based Pim inhibitors: A template-based approach.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4MBL
 
 | | Discovery of Pyrazolo[1,5a]pyrimidine-based Pim1 Inhibitors | | Descriptor: | (1R,2R)-N-[3-(naphthalen-2-yl)pyrazolo[1,5-a]pyrimidin-5-yl]cyclohexane-1,2-diamine, Serine/threonine-protein kinase pim-1 | | Authors: | Azevedo, R, Fischmann, T.O. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of pyrazolo[1,5-a]pyrimidine-based Pim inhibitors: A template-based approach.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7UP2
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (2M)-4'-methyl-2-(2H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UP1
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (2M)-4'-methyl-2-(2H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, CADMIUM ION, Metallo beta-lactamase, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UP3
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (3P)-4-[4-(hydroxymethyl)phenyl]-3-(2H-tetrazol-5-yl)pyridine-2-sulfonamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CADMIUM ION, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UOX
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (2M)-4'-(hydroxymethyl)-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-ol, ACETATE ION, CADMIUM ION, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UOY
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (6P)-4-amino-6-(2H-tetrazol-5-yl)benzene-1,3-disulfonamide, CADMIUM ION, Metallo beta-lactamase, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
