4ESW
 
 | | Crystal structure of C. albicans Thi5 H66G mutant | | Descriptor: | CITRIC ACID, Pyrimidine biosynthesis enzyme THI13 | | Authors: | Fenwick, M.K, Huang, S, Zhang, Y, Lai, R, Hazra, A, Rajashankar, K, Philmus, B, Kinsland, C, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-04-23 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Thiamin pyrimidine biosynthesis in Candida albicans : a remarkable reaction between histidine and pyridoxal phosphate.
J.Am.Chem.Soc., 134, 2012
|
|
5UFM
 
 | | Crystal structure of Burkholderia thailandensis 1,6-didemethyltoxoflavin-N1-methyltransferase with bound 1,6-didemethyltoxoflavin and S-adenosylhomocysteine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Methyltransferase domain protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Fenwick, M.K, Ealick, S.E, Almabruk, K.H, Begley, T.P, Philmus, B. | | Deposit date: | 2017-01-05 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Biochemical Characterization and Structural Basis of Reactivity and Regioselectivity Differences between Burkholderia thailandensis and Burkholderia glumae 1,6-Didesmethyltoxoflavin N-Methyltransferase.
Biochemistry, 56, 2017
|
|
5UFN
 
 | | Crystal structure of Burkholderia thailandensis 1,6-didemethyltoxoflavin-N1-methyltransferase with bound S-adenosylhomocysteine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Methyltransferase domain protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Fenwick, M.K, Ealick, S.E, Almabruk, K.H, Begley, T.P, Philmus, B. | | Deposit date: | 2017-01-05 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Biochemical Characterization and Structural Basis of Reactivity and Regioselectivity Differences between Burkholderia thailandensis and Burkholderia glumae 1,6-Didesmethyltoxoflavin N-Methyltransferase.
Biochemistry, 56, 2017
|
|
8FBQ
 
 | | Crystal structure of Plasmodium vivax glycylpeptide N-tetradecanoyltransferase (N-myristoyltransferase, NMT) bound to myristoyl-CoA and inhibitor 12b | | Descriptor: | 1-[(3M)-3-{3-[2-(1,3,5-trimethyl-1H-pyrazol-4-yl)ethoxy]pyridin-2-yl}phenyl]piperazine, ACETATE ION, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Staker, B.L, Lovell, S.W, Phan, I.Q, Early, J, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-11-29 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of potent and selective N-myristoyltransferase inhibitors of Plasmodium vivax liver stage hypnozoites and schizonts.
Nat Commun, 14, 2023
|
|
6Q2P
 
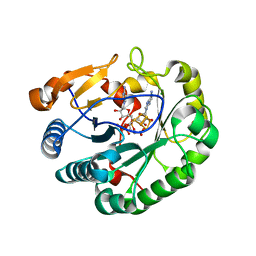 | | Crystal structure of mouse viperin bound to cytidine triphosphate and S-adenosylhomocysteine | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Fenwick, M.K, Dong, M, Lin, H, Ealick, S.E. | | Deposit date: | 2019-08-08 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | Structural Basis of the Substrate Selectivity of Viperin.
Biochemistry, 59, 2020
|
|
6Q2Q
 
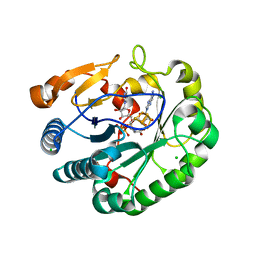 | | Crystal structure of mouse viperin bound to uridine triphosphate and S-adenosylhomocysteine | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Fenwick, M.K, Dong, M, Lin, H, Ealick, S.E. | | Deposit date: | 2019-08-08 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural Basis of the Substrate Selectivity of Viperin.
Biochemistry, 59, 2020
|
|
6Q2E
 
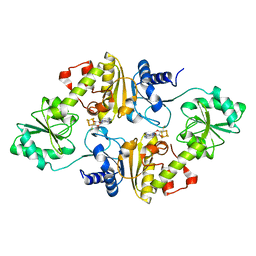 | | Crystal structure of Methanobrevibacter smithii Dph2 bound to 5'-methylthioadenosine | | Descriptor: | 2-(3-amino-3-carboxypropyl)histidine synthase, 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Dong, M, Lin, H, Ealick, S.E. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | The Crystal Structure of Dph2 in Complex with Elongation Factor 2 Reveals the Structural Basis for the First Step of Diphthamide Biosynthesis.
Biochemistry, 58, 2019
|
|
6Q2D
 
 | | Crystal structure of Methanobrevibacter smithii Dph2 in complex with Methanobrevibacter smithii elongation factor 2 | | Descriptor: | 2-(3-amino-3-carboxypropyl)histidine synthase, Elongation factor 2, IRON/SULFUR CLUSTER | | Authors: | Fenwick, M.K, Dong, M, Lin, H, Ealick, S.E. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The Crystal Structure of Dph2 in Complex with Elongation Factor 2 Reveals the Structural Basis for the First Step of Diphthamide Biosynthesis.
Biochemistry, 58, 2019
|
|
6U44
 
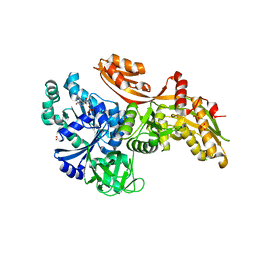 | |
5VSL
 
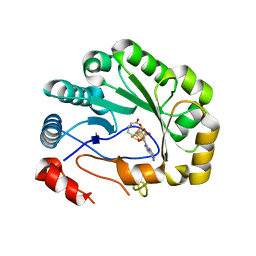 | | Crystal structure of viperin with bound [4Fe-4S] cluster and S-adenosylhomocysteine (SAH) | | Descriptor: | IRON/SULFUR CLUSTER, Radical S-adenosyl methionine domain-containing protein 2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Fenwick, M.K, Li, Y, Cresswell, P, Modis, Y, Ealick, S.E. | | Deposit date: | 2017-05-11 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Structural studies of viperin, an antiviral radical SAM enzyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4S26
 
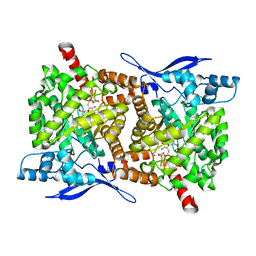 | | Crystal structure of Arabidopsis thaliana ThiC with bound imidazole ribonucleotide, S-adenosylhomocysteine, Fe4S4 cluster and Zn (monoclinic crystal form) | | Descriptor: | 1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
5VSM
 
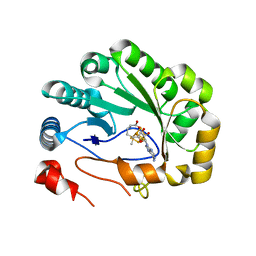 | | Crystal structure of viperin with bound [4Fe-4S] cluster, 5'-deoxyadenosine, and L-methionine | | Descriptor: | 5'-DEOXYADENOSINE, IRON/SULFUR CLUSTER, METHIONINE, ... | | Authors: | Fenwick, M.K, Li, Y, Cresswell, P, Modis, Y, Ealick, S.E. | | Deposit date: | 2017-05-11 | | Release date: | 2017-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of viperin, an antiviral radical SAM enzyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6BXO
 
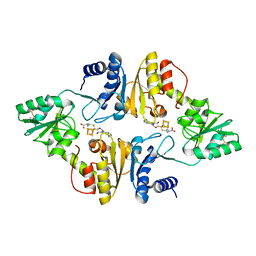 | | Crystal structure of Candidatus Methanoperedens nitroreducens Dph2 with 4Fe-4S cluster and SAH | | Descriptor: | Diphthamide biosynthesis enzyme Dph2, IRON/SULFUR CLUSTER, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Fenwick, M.K, Torelli, A.T, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
6BXN
 
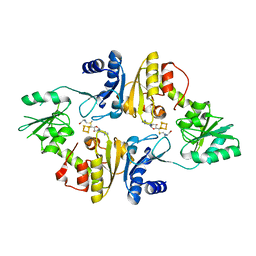 | | Crystal structure of Candidatus Methanoperedens nitroreducens Dph2 with 4Fe-4S cluster and SAM | | Descriptor: | Diphthamide biosynthesis enzyme Dph2, IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE | | Authors: | Fenwick, M.K, Torelli, A.T, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
6BXM
 
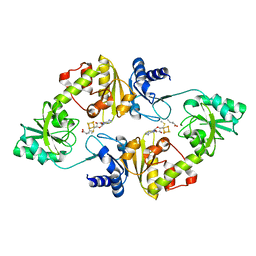 | | Crystal structure of Candidatus Methanoperedens nitroreducens Dph2 with 4Fe-4S cluster and SAM/cleaved SAM | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, ALPHA-AMINOBUTYRIC ACID, Diphthamide biosynthesis enzyme Dph2, ... | | Authors: | Fenwick, M.K, Torelli, A.T, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
6U45
 
 | |
6U43
 
 | |
3PKV
 
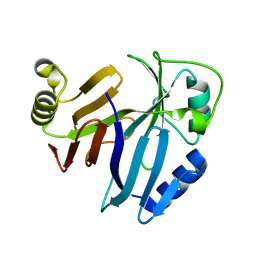 | |
3PKX
 
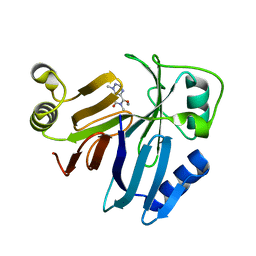 | | Crystal Structure of Toxoflavin Lyase (TflA) bound to Mn(II) and Toxoflavin | | Descriptor: | 1,6-dimethylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione, MANGANESE (II) ION, Toxoflavin lyase (TflA) | | Authors: | Fenwick, M.K, Philmus, B, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-11-12 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Toxoflavin lyase requires a novel 1-his-2-carboxylate facial triad .
Biochemistry, 50, 2011
|
|
3PKW
 
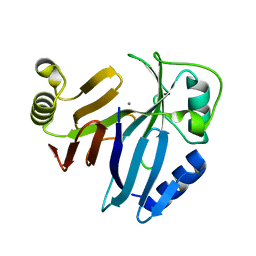 | | Crystal Structure of Toxoflavin Lyase (TflA) bound to Mn(II) | | Descriptor: | MANGANESE (II) ION, Toxoflavin lyase (TflA) | | Authors: | Fenwick, M.K, Philmus, B, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-11-12 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toxoflavin lyase requires a novel 1-his-2-carboxylate facial triad .
Biochemistry, 50, 2011
|
|
4S29
 
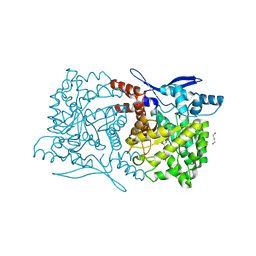 | | Crystal structure of Arabidopsis thaliana ThiC with bound imidazole ribonucleotide and Fe | | Descriptor: | 1,4-BUTANEDIOL, 1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole, FE (II) ION, ... | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.382 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
4S25
 
 | | Crystal structure of Arabidopsis thaliana ThiC with bound imidazole ribonucleotide, S-adenosylhomocysteine, Fe4S4 cluster and Zn (trigonal crystal form) | | Descriptor: | 1,4-BUTANEDIOL, 1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
4S2A
 
 | | Crystal structure of Caulobacter crescentus ThiC with Fe4S4 cluster at remote site (holo form) | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHATE ION, Phosphomethylpyrimidine synthase | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
4S27
 
 | | Crystal structure of Arabidopsis thaliana ThiC with bound aminoimidazole ribonucleotide, 5'-deoxyadenosine, L-methionine, Fe4S4 cluster and Fe | | Descriptor: | 1,4-BUTANEDIOL, 5'-DEOXYADENOSINE, 5-AMINOIMIDAZOLE RIBONUCLEOTIDE, ... | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
4S28
 
 | | Crystal structure of Arabidopsis thaliana ThiC with bound aminoimidazole ribonucleotide, S-adenosylhomocysteine, Fe4S4 cluster and Fe | | Descriptor: | 1,4-BUTANEDIOL, 5-AMINOIMIDAZOLE RIBONUCLEOTIDE, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
