1HOV
 
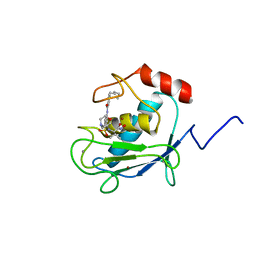 | | SOLUTION STRUCTURE OF A CATALYTIC DOMAIN OF MMP-2 COMPLEXED WITH SC-74020 | | Descriptor: | CALCIUM ION, MATRIX METALLOPROTEINASE-2, N-{4-[(1-HYDROXYCARBAMOYL-2-METHYL-PROPYL)-(2-MORPHOLIN-4-YL-ETHYL)-SULFAMOYL]-4-PENTYL-BENZAMIDE, ... | | Authors: | Feng, Y, Likos, J.J, Zhu, L, Woodward, H, Munie, G, McDonald, J.J, Stevens, A.M, Howard, C.P, De Crescenzo, G.A, Welsch, D, Shieh, H.-S, Stallings, W.C. | | Deposit date: | 2000-12-11 | | Release date: | 2001-12-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the catalytic domain of matrix metalloproteinase-2 complexed with a hydroxamic acid inhibitor
Biochim.Biophys.Acta, 1598, 2002
|
|
6INW
 
 | | A Pericyclic Reaction enzyme | | Descriptor: | O-methyltransferase lepI, S-ADENOSYLMETHIONINE | | Authors: | Feng, Y, Chang, M, Wang, H, Liu, Z, Zhou, Y. | | Deposit date: | 2018-10-28 | | Release date: | 2019-07-03 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Crystal structure of the multifunctional SAM-dependent enzyme LepI provides insights into its catalytic mechanism.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
6IGM
 
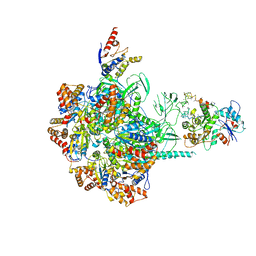 | | Cryo-EM Structure of Human SRCAP Complex | | Descriptor: | Actin-related protein 6, Helicase SRCAP, RuvB-like 1, ... | | Authors: | Feng, Y, Tian, Y, Wu, Z, Xu, Y. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of human SRCAP complex.
Cell Res., 28, 2018
|
|
1JLI
 
 | |
2MK5
 
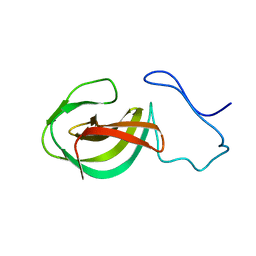 | | Solution structure of a protein domain | | Descriptor: | Endolysin | | Authors: | Feng, Y, Gu, J. | | Deposit date: | 2014-01-24 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and biochemical characterization reveals LysGH15 as an unprecedented "EF-hand-like" calcium-binding phage lysin.
Plos Pathog., 10, 2014
|
|
5YIM
 
 | | Structure of a Legionella effector | | Descriptor: | SdeA | | Authors: | Feng, Y, Dong, Y, Wang, W. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.394 Å) | | Cite: | Structural basis of ubiquitin modification by the Legionella effector SdeA.
Nature, 557, 2018
|
|
5YIK
 
 | |
5YIJ
 
 | | Structure of a Legionella effector with substrates | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, SdeA, Ubiquitin | | Authors: | Feng, Y, Mu, Y, Wang, H. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structural basis of ubiquitin modification by the Legionella effector SdeA.
Nature, 557, 2018
|
|
1Z7P
 
 | | Solution structure of reduced glutaredoxin C1 from Populus tremula x tremuloides | | Descriptor: | glutaredoxin | | Authors: | Feng, Y, Zhong, N, Rouhier, N, Jacquot, J.P, Xia, B. | | Deposit date: | 2005-03-26 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into Poplar Glutaredoxin C1 with a Bridging Iron-Sulfur Cluster at the Active Site
Biochemistry, 45, 2006
|
|
8H2J
 
 | | Structure of Acb2 complexed with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, p26 | | Authors: | Feng, Y, Cao, X.L. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacteriophages inhibit and evade cGAS-like immune function in bacteria.
Cell, 186, 2023
|
|
8H2X
 
 | | Structure of Acb2 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, p26 | | Authors: | Feng, Y, Cao, X.L. | | Deposit date: | 2022-10-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Bacteriophages inhibit and evade cGAS-like immune function in bacteria.
Cell, 186, 2023
|
|
8H39
 
 | | Structure of Acb2 complexed with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, 1,2-ETHANEDIOL, p26 | | Authors: | Feng, Y, Cao, X.L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Bacteriophages inhibit and evade cGAS-like immune function in bacteria.
Cell, 186, 2023
|
|
3CVA
 
 | | Human Bcl-xL containing a Trp to Ala mutation at position 137 | | Descriptor: | Apoptosis regulator Bcl-X | | Authors: | Feng, Y, Zhang, L, Hu, T, Shen, X, Chen, K, Jiang, H, Liu, D. | | Deposit date: | 2008-04-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A conserved hydrophobic core at Bcl-x(L) mediates its structural stability and binding affinity with BH3-domain peptide of pro-apoptotic protein
Arch.Biochem.Biophys., 484, 2009
|
|
4ZH2
 
 | |
4ZH3
 
 | |
4ZH4
 
 | | Crystal structure of Escherichia coli RNA polymerase in complex with CBRP18 | | Descriptor: | 5-(4-fluorophenyl)-4-[4-fluoro-3-(trifluoromethyl)phenyl]-1H-pyrazole, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Feng, Y, Ebright, R.H. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.993 Å) | | Cite: | Structural Basis of Transcription Inhibition by CBR Hydroxamidines and CBR Pyrazoles.
Structure, 23, 2015
|
|
6A9Y
 
 | | The crystal structure of Mu homology domain of SGIP1 | | Descriptor: | SH3-containing GRB2-like protein 3-interacting protein 1 | | Authors: | Feng, Y, Liu, X. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SGIP1 dimerizes via intermolecular disulfide bond in mu HD domain during cellular endocytosis.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
1Z7R
 
 | | Solution Structure of reduced glutaredoxin C1 from Populus tremula x tremuloides | | Descriptor: | glutaredoxin | | Authors: | Feng, Y, Zhong, N, Rouhier, N, Jacquot, J.P, Xia, B. | | Deposit date: | 2005-03-26 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into Poplar Glutaredoxin C1 with a Bridging Iron-Sulfur Cluster at the Active Site
Biochemistry, 45, 2006
|
|
6SXN
 
 | | Crystal structure of P212121 apo form of CrtE | | Descriptor: | Geranylgeranyl pyrophosphate synthase | | Authors: | Feng, Y, Morgan, R.M.L, Nixon, P.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase (CrtE) Involved in Cyanobacterial Terpenoid Biosynthesis.
Front Plant Sci, 11, 2020
|
|
6SXL
 
 | | Crystal structure of CrtE | | Descriptor: | Geranylgeranyl pyrophosphate synthase, PHOSPHATE ION | | Authors: | Feng, Y, Morgan, R.M.L, Nixon, P.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase (CrtE) Involved in Cyanobacterial Terpenoid Biosynthesis.
Front Plant Sci, 11, 2020
|
|
3KXT
 
 | | Crystal structure of Sulfolobus Cren7-dsDNA complex | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3', Chromatin protein Cren7 | | Authors: | Feng, Y, Wang, J. | | Deposit date: | 2009-12-03 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Crystal structure of the crenarchaeal conserved chromatin protein Cren7 and double-stranded DNA complex
Protein Sci., 19, 2010
|
|
5XVM
 
 | |
4JND
 
 | | Structure of a C.elegans sex determining protein | | Descriptor: | Ca(2+)/calmodulin-dependent protein kinase phosphatase, MAGNESIUM ION | | Authors: | Feng, Y, Zhang, Y, Ge, J, Yang, M. | | Deposit date: | 2013-03-15 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Structural insight into Caenorhabditis elegans sex-determining protein FEM-2.
J.Biol.Chem., 288, 2013
|
|
2LEO
 
 | |
5X38
 
 | |
