4PWM
 
 | | Crystal structure of Dickerson Drew Dodecamer with 5-carboxycytosine | | Descriptor: | 5'-[CGCGAATT(5CC)GCG]-3' | | Authors: | Szulik, M.W, Pallan, P, Banerjee, S, Voehler, M, Egli, M, Stone, M.P. | | Deposit date: | 2014-03-20 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Differential stabilities and sequence-dependent base pair opening dynamics of watson-crick base pairs with 5-hydroxymethylcytosine, 5-formylcytosine, or 5-carboxylcytosine.
Biochemistry, 54, 2015
|
|
7JJE
 
 | |
7JJF
 
 | | Sarcin-ricin loop with modified residue. | | Descriptor: | MAGNESIUM ION, RNA/DNA (27-mer) | | Authors: | Harp, J.M, Pallan, P.S, Egli, M. | | Deposit date: | 2020-07-25 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Incorporating a Thiophosphate Modification into a Common RNA Tetraloop Motif Causes an Unanticipated Stability Boost.
Biochemistry, 59, 2020
|
|
7JJD
 
 | |
7JY2
 
 | | Z-DNA joint X-ray/Neutron | | Descriptor: | Chains: A,B | | Authors: | Harp, J.M, Coates, L, Egli, M. | | Deposit date: | 2020-08-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | Cite: | Water structure around a left-handed Z-DNA fragment analyzed by cryo neutron crystallography.
Nucleic Acids Res., 49, 2021
|
|
5JUM
 
 | | Crystal Structure of Human DNA Polymerase Eta Inserting dCTP Opposite N-(2'-deoxyguanosin-8- yl)-3-aminobenzanthrone (C8-dG-ABA) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Patra, A, Politica, D.A, Stone, M.P, Egli, M. | | Deposit date: | 2016-05-10 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of Error-Free Bypass of the Environmental Carcinogen N-(2'-Deoxyguanosin-8-yl)-3-aminobenzanthrone Adduct by Human DNA Polymerase eta.
Chembiochem, 17, 2016
|
|
3OZ5
 
 | | S-Methyl Carbocyclic LNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(UMX)P*AP*CP*GP*C)-3'), SPERMINE | | Authors: | Seth, P.R, Allerson, C.A, Berdeja, A, Siwkowski, A, Pallan, P.S, Gaus, H, Prakash, T.P, Watt, A.T, Egli, M, Swayze, E.E. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | An exocyclic methylene group acts as a bioisostere of the 2'-oxygen atom in LNA.
J.Am.Chem.Soc., 132, 2010
|
|
3OZ3
 
 | | Vinyl Carbocyclic LNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*(UVX)P*AP*CP*GP*C)-3') | | Authors: | Seth, P.R, Allerson, C.A, Berdeja, A, Siwkowski, A, Pallan, P.S, Gaus, H, Prakash, T.P, Watt, A.T, Egli, M, Swayze, E.E. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | An exocyclic methylene group acts as a bioisostere of the 2'-oxygen atom in LNA.
J.Am.Chem.Soc., 132, 2010
|
|
3OZ4
 
 | | R-Methyl Carbocyclic LNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(URX)P*AP*CP*GP*C)-3') | | Authors: | Seth, P.R, Allerson, C.A, Berdeja, A, Siwkowski, A, Pallan, P.S, Gaus, H, Prakash, T.P, Watt, A.T, Egli, M, Swayze, E.E. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | An exocyclic methylene group acts as a bioisostere of the 2'-oxygen atom in LNA.
J.Am.Chem.Soc., 132, 2010
|
|
6XHC
 
 | | Structure of glycinyl 5'-O-adenosine phosphoramidate | | Descriptor: | 2-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]amino]ethanoic acid, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XHE
 
 | | Structure of beta-prolinyl 5'-O-adenosine phosphoramidate | | Descriptor: | 5'-O-[(S)-[(3S)-3-carboxypyrrolidin-1-yl](hydroxy)phosphoryl]adenosine, ADENOSINE MONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XHD
 
 | | Structure of Prolinyl-5'-O-adenosine phosphoramidate | | Descriptor: | (2~{S})-1-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]pyrrolidine-2-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XHF
 
 | | Structure of Phenylalanyl-5'-O-adenosine phosphoramidate | | Descriptor: | (2~{S})-2-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]amino]-3-phenyl-propanoic acid, CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5L1I
 
 | |
5L1L
 
 | | PostInsertion complex of Human DNA Polymerase Eta bypassing an O6-Methyl-2'-deoxyguanosine : dT site | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*GP*(6OG)P*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Mechanisms of Insertion of dCTP and dTTP Opposite the DNA Lesion O6-Methyl-2'-deoxyguanosine by Human DNA Polymerase eta.
J.Biol.Chem., 291, 2016
|
|
5L1K
 
 | | PostInsertion complex of Human DNA Polymerase Eta bypassing an O6-Methyl-2'-deoxyguanosine : dC site | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*C)-3'), DNA (5'-D(*CP*AP*TP*GP*(6OG)P*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Mechanisms of Insertion of dCTP and dTTP Opposite the DNA Lesion O6-Methyl-2'-deoxyguanosine by Human DNA Polymerase eta.
J.Biol.Chem., 291, 2016
|
|
5L1J
 
 | |
3Q61
 
 | | 3'-Fluoro Hexitol Nucleic Acid DNA Structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(F3H)P*AP*CP*GP*C)-3') | | Authors: | Seth, P.R, Allerson, C.R, Prakash, T.P, Siwkowski, A, Berdeja, A, Yu, J, Pallan, P.S, Watt, A.T, Gaus, H, Bhat, B, Egli, M, Swayze, E.E. | | Deposit date: | 2010-12-30 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Synthesis, improved antisense activity and structural rationale for the divergent RNA affinities of 3'-fluoro hexitol nucleic acid (FHNA and Ara-FHNA) modified oligonucleotides.
J.Am.Chem.Soc., 133, 2011
|
|
4QC7
 
 | | Dodecamer structure of 5-formylcytosine containing DNA | | Descriptor: | short DNA strands | | Authors: | Szulik, M.W, Pallan, P, Egli, M, Stone, M.P. | | Deposit date: | 2014-05-09 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential stabilities and sequence-dependent base pair opening dynamics of watson-crick base pairs with 5-hydroxymethylcytosine, 5-formylcytosine, or 5-carboxylcytosine.
Biochemistry, 54, 2015
|
|
3S1A
 
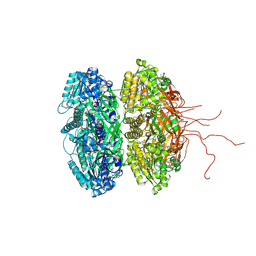 | | Crystal structure of the phosphorylation-site double mutant S431E/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Williams, D.W, Rossi, G, Weigand, S, Mori, T, Johnson, C.H, Stewart, P.L, Egli, M. | | Deposit date: | 2011-05-14 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Combined SAXS/EM Based Models of the S. elongatus Post-Translational Circadian Oscillator and its Interactions with the Output His-Kinase SasA.
Plos One, 6, 2011
|
|
6B82
 
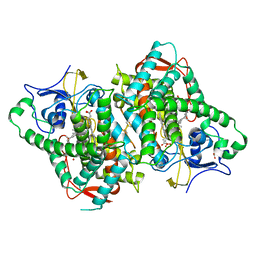 | | Zebra Fish CYP-450 17A1 Mutant Abiraterone Complex | | Descriptor: | ACETATE ION, Abiraterone, CHLORIDE ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-10-05 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Inherent steroid 17 alpha ,20-lyase activity in defunct cytochrome P450 17A enzymes.
J. Biol. Chem., 293, 2018
|
|
3UKC
 
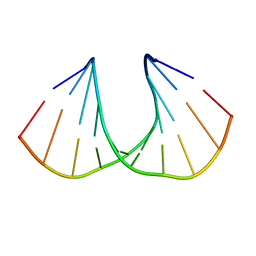 | | (S)-cEt-BNA decamer structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(1TL)P*AP*CP*GP*C)-3') | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure and nuclease resistance of 2',4'-constrained 2'-O-methoxyethyl (cMOE) and 2'-O-ethyl (cEt) modified DNAs.
Chem.Commun.(Camb.), 48, 2012
|
|
3UKE
 
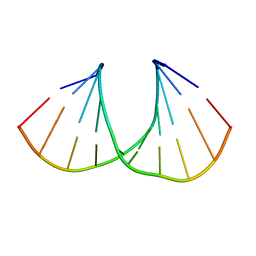 | | (S)-cMOE-BNA decamer structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(CSM)P*AP*CP*GP*C)-3') | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure and nuclease resistance of 2',4'-constrained 2'-O-methoxyethyl (cMOE) and 2'-O-ethyl (cEt) modified DNAs.
Chem.Commun.(Camb.), 48, 2012
|
|
3UKB
 
 | | (R)-cEt-BNA decamer structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(RCE)P*AP*CP*GP*C)-3') | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure and nuclease resistance of 2',4'-constrained 2'-O-methoxyethyl (cMOE) and 2'-O-ethyl (cEt) modified DNAs.
Chem.Commun.(Camb.), 48, 2012
|
|
3V06
 
 | | Crystal structure of S-6'-Me-3'-fluoro hexitol nucleic acid | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(F5H)P*AP*CP*GP*C)-3'), STRONTIUM ION | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2011-12-07 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Insights from crystal structures into the opposite effects on RNA affinity caused by the s- and R-6'-methyl backbone modifications of 3'-fluoro hexitol nucleic Acid.
Biochemistry, 51, 2012
|
|
