3M1C
 
 | |
3MIE
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound azide obtained by soaking (50mM NaN3) | | Descriptor: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3MLJ
 
 | | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound carbon monooxide (CO) | | Descriptor: | ACETATE ION, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Prigge, S.T, Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Differential reactivity between two copper sites in peptidylglycine alpha-hydroxylating monooxygenase
J.Am.Chem.Soc., 132, 2010
|
|
3MIG
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound nitrite, obtained in the presence of substrate | | Descriptor: | COPPER (II) ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3NWD
 
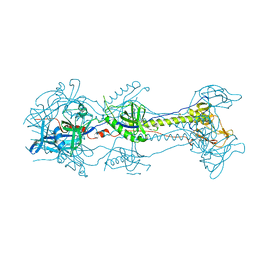 | | Glycoprotein B from Herpes simplex virus type 1, Y179S mutant, low-pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Stampfer, S.D, Lou, H, Cohen, G.H, Eisenberg, R.J, Heldwein, E.E. | | Deposit date: | 2010-07-09 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8803 Å) | | Cite: | Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1.
J.Virol., 84, 2010
|
|
3MLL
 
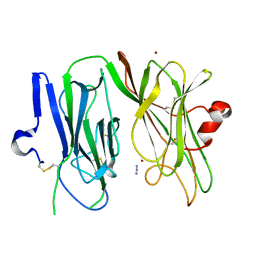 | | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound azide | | Descriptor: | AZIDE ION, COPPER (II) ION, NICKEL (II) ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Differential reactivity between two copper sites in peptidylglycine alpha-hydroxylating monooxygenase
J.Am.Chem.Soc., 132, 2010
|
|
3MIB
 
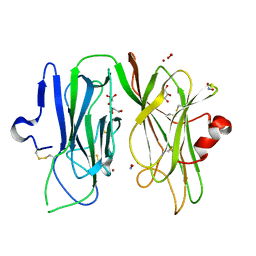 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound nitrite | | Descriptor: | COPPER (II) ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3OJE
 
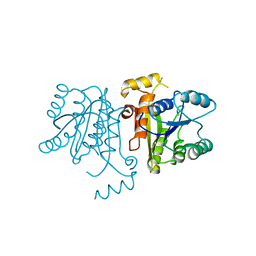 | | Crystal Structure of the Bacillus cereus Enoyl-Acyl Carrier Protein Reductase (Apo form) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase (FabL) (NADPH) | | Authors: | Kim, S.J, Ha, B.H, Kim, K.H, Hong, S.K, Suh, S.W, Kim, E.E. | | Deposit date: | 2010-08-22 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Dimeric and tetrameric forms of enoyl-acyl carrier protein reductase from Bacillus cereus
Biochem.Biophys.Res.Commun., 400, 2010
|
|
3OIC
 
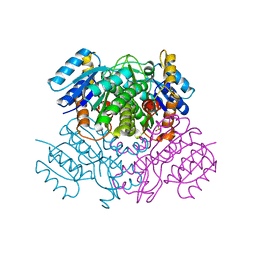 | | Crystal Structure of Enoyl-ACP Reductases III (FabL) from B. subtilis (apo form) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH], SULFATE ION | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
3OZ3
 
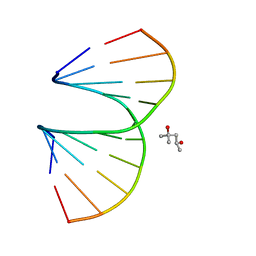 | | Vinyl Carbocyclic LNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*(UVX)P*AP*CP*GP*C)-3') | | Authors: | Seth, P.R, Allerson, C.A, Berdeja, A, Siwkowski, A, Pallan, P.S, Gaus, H, Prakash, T.P, Watt, A.T, Egli, M, Swayze, E.E. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | An exocyclic methylene group acts as a bioisostere of the 2'-oxygen atom in LNA.
J.Am.Chem.Soc., 132, 2010
|
|
3OIF
 
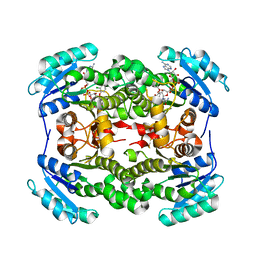 | | Crystal Structure of Enoyl-ACP Reductases I (FabI) from B. subtilis (complex with NAD and TCL) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
3MIC
 
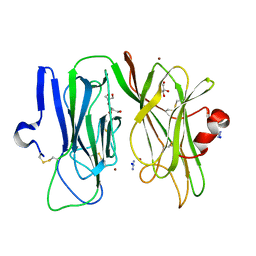 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound azide obtained by co-crystallization | | Descriptor: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3OIG
 
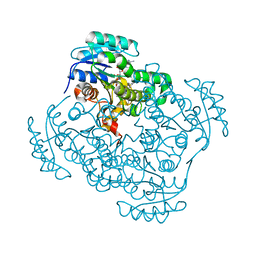 | | Crystal Structure of Enoyl-ACP Reductases I (FabI) from B. subtilis (complex with NAD and INH) | | Descriptor: | (2E)-N-[(1,2-dimethyl-1H-indol-3-yl)methyl]-N-methyl-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)prop-2-enamide, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
3OJF
 
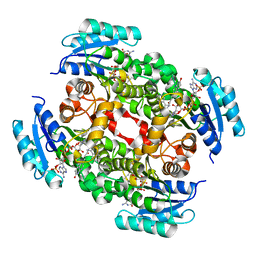 | | Crystal Structure of the Bacillus cereus Enoyl-Acyl Carrier Protein Reductase with NADP+ and indole naphthyridinone (Complex form) | | Descriptor: | (2E)-N-[(1,2-dimethyl-1H-indol-3-yl)methyl]-N-methyl-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)prop-2-enamide, Enoyl-[acyl-carrier-protein] reductase (FabL) (NADPH), NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kim, S.J, Ha, B.H, Kim, K.H, Hong, S.K, Suh, S.W, Kim, E.E. | | Deposit date: | 2010-08-22 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dimeric and tetrameric forms of enoyl-acyl carrier protein reductase from Bacillus cereus
Biochem.Biophys.Res.Commun., 400, 2010
|
|
3OZ5
 
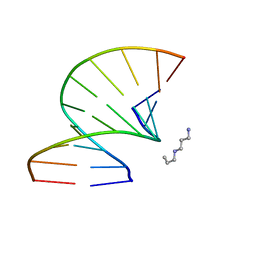 | | S-Methyl Carbocyclic LNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(UMX)P*AP*CP*GP*C)-3'), SPERMINE | | Authors: | Seth, P.R, Allerson, C.A, Berdeja, A, Siwkowski, A, Pallan, P.S, Gaus, H, Prakash, T.P, Watt, A.T, Egli, M, Swayze, E.E. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | An exocyclic methylene group acts as a bioisostere of the 2'-oxygen atom in LNA.
J.Am.Chem.Soc., 132, 2010
|
|
3OQC
 
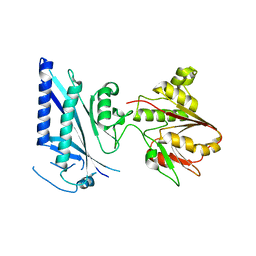 | |
3OID
 
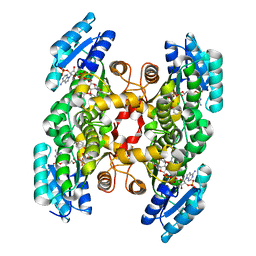 | | Crystal Structure of Enoyl-ACP Reductases III (FabL) from B. subtilis (complex with NADP and TCL) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRICLOSAN | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
3OZ4
 
 | | R-Methyl Carbocyclic LNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(URX)P*AP*CP*GP*C)-3') | | Authors: | Seth, P.R, Allerson, C.A, Berdeja, A, Siwkowski, A, Pallan, P.S, Gaus, H, Prakash, T.P, Watt, A.T, Egli, M, Swayze, E.E. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | An exocyclic methylene group acts as a bioisostere of the 2'-oxygen atom in LNA.
J.Am.Chem.Soc., 132, 2010
|
|
1SLS
 
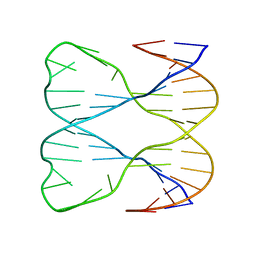 | | IMMOBILE SLIPPED-LOOP STRUCTURE (SLS) OF DNA HOMODIMER IN SOLUTION, NMR, 9 STRUCTURES | | Descriptor: | OLIGODEOXYRIBONUCLEOTIDE | | Authors: | Ulyanov, N.B, Ivanov, V.I, Minyat, E.E, Khomyakova, E.B, Petrova, M.V, Lesiak, K, James, T.L. | | Deposit date: | 1997-09-30 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A pseudosquare knot structure of DNA in solution.
Biochemistry, 37, 1998
|
|
1SND
 
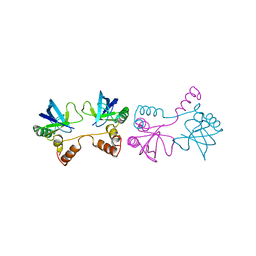 | | STAPHYLOCOCCAL NUCLEASE DIMER CONTAINING A DELETION OF RESIDUES 114-119 COMPLEXED WITH CALCIUM CHLORIDE AND THE COMPETITIVE INHIBITOR DEOXYTHYMIDINE-3',5'-DIPHOSPHATE | | Descriptor: | STAPHYLOCOCCAL NUCLEASE DIMER | | Authors: | Green, S.M, Gittis, A.G, Meeker, A.K, Lattman, E.E. | | Deposit date: | 1996-08-23 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | One-step evolution of a dimer from a monomeric protein.
Nat.Struct.Biol., 2, 1995
|
|
1STG
 
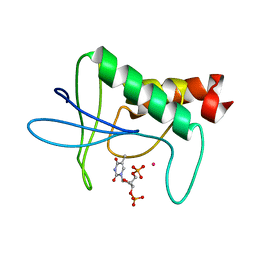 | | TWO DISTINCTLY DIFFERENT METAL BINDING MODES ARE SEEN IN X-RAY CRYSTAL STRUCTURES OF STAPHYLOCOCCAL NUCLEASE-COBALT(II)-NUCLEOTIDE COMPLEXES | | Descriptor: | COBALT (II) ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Loll, P.J, Quirk, S, Lattman, E.E. | | Deposit date: | 1994-10-27 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray crystal structures of staphylococcal nuclease complexed with the competitive inhibitor cobalt(II) and nucleotide.
Biochemistry, 34, 1995
|
|
1STH
 
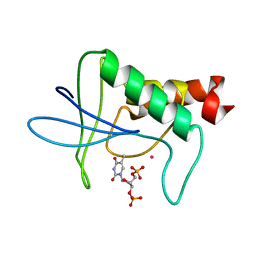 | | TWO DISTINCTLY DIFFERENT METAL BINDING MODES ARE SEEN IN X-RAY CRYSTAL STRUCTURES OF STAPHYLOCOCCAL NUCLEASE-COBALT(II)-NUCLEOTIDE COMPLEXES | | Descriptor: | COBALT (II) ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Loll, P.J, Quirk, S, Lattman, E.E. | | Deposit date: | 1994-10-27 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystal structures of staphylococcal nuclease complexed with the competitive inhibitor cobalt(II) and nucleotide.
Biochemistry, 34, 1995
|
|
1TCO
 
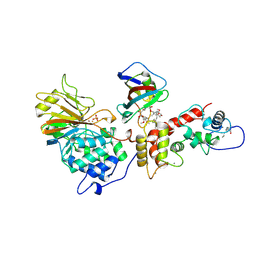 | | TERNARY COMPLEX OF A CALCINEURIN A FRAGMENT, CALCINEURIN B, FKBP12 AND THE IMMUNOSUPPRESSANT DRUG FK506 (TACROLIMUS) | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, FE (III) ION, ... | | Authors: | Griffith, J.P, Kim, J.L, Kim, E.E, Sintchak, M.D, Thomson, J.A, Fitzgibbon, M.J, Fleming, M.A, Caron, P.R, Hsiao, K, Navia, M.A. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex.
Cell(Cambridge,Mass.), 82, 1995
|
|
1TQO
 
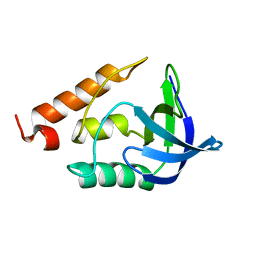 | |
1RRE
 
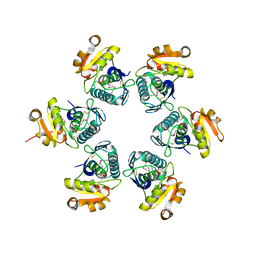 | | Crystal structure of E.coli Lon proteolytic domain | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Rasulova, F, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
