9AZ7
 
 | | Chloride Sites in Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, CHLORIDE ION, Photoactive yellow protein | | Authors: | Dyda, F, Schotte, F, Anfinrud, P, Cho, H.S. | | Deposit date: | 2024-03-10 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Watching a signaling protein function: What has been learned over four decades of time-resolved studies of photoactive yellow protein.
Struct Dyn., 11, 2024
|
|
9AZ9
 
 | | Chloride Sites in Photoactive Yellow Protein (Chloride-Free Reference Structure) | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Dyda, F, Schotte, F, Anfinrud, P, Cho, H.S. | | Deposit date: | 2024-03-10 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Watching a signaling protein function: What has been learned over four decades of time-resolved studies of photoactive yellow protein.
Struct Dyn., 11, 2024
|
|
6B67
 
 | | Human PP2Calpha (PPM1A) complexed with cyclic peptide c(MpSIpYVA) | | Descriptor: | CALCIUM ION, Protein phosphatase 1A, cyclic peptide c(MpSIpYVA) | | Authors: | Dyda, F, Kosek, D. | | Deposit date: | 2017-10-01 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A trapped human PPM1A-phosphopeptide complex reveals structural features critical for regulation of PPM protein phosphatase activity.
J. Biol. Chem., 293, 2018
|
|
6OPM
 
 | | Casposase bound to integration product | | Descriptor: | CALCIUM ION, CRISPR-associated endonuclease Cas1, DNA 21-mer, ... | | Authors: | Dyda, F, Hickman, A.B, Kailasan, S. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-12 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Casposase structure and the mechanistic link between DNA transposition and spacer acquisition by CRISPR-Cas.
Elife, 9, 2020
|
|
1UUT
 
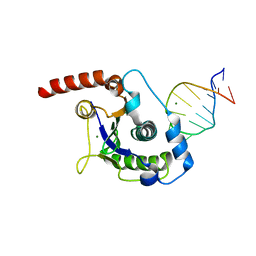 | | The Nuclease Domain of Adeno-Associated Virus Rep Complexed with the RBE' Stemloop of the Viral Inverted Terminal Repeat | | Descriptor: | 5'-D(*CP*AP*GP*CP*TP*CP*TP*TP*TP*GP *AP*GP*CP*TP*G)-3', CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Dyda, F, Hickman, A.B, Ronning, D.R, Perez, Z.N, Kotin, R.M. | | Deposit date: | 2004-01-10 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Nuclease Domain of Adeno-Associated Virus Rep Coordinates Replication Initiation Using Two Distinct DNA Recognition Interfaces
Mol.Cell, 13, 2004
|
|
6DWZ
 
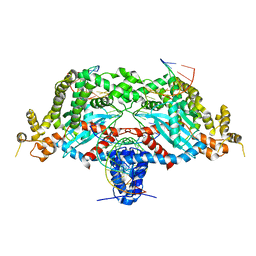 | | Hermes transposase deletion dimer complex with (C/G) DNA | | Descriptor: | DNA (26-MER), DNA (5'-D(*GP*AP*GP*AP*AP*CP*AP*AP*CP*AP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*C)-3'), ... | | Authors: | Dyda, F, Hickman, A.B. | | Deposit date: | 2018-06-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase.
Nucleic Acids Res., 46, 2018
|
|
6DWW
 
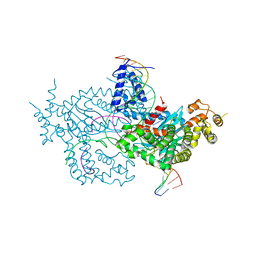 | | Hermes transposase deletion dimer complex with (A/T) DNA and Mn2+ | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*GP*AP*GP*AP*AP*CP*AP*AP*CP*AP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*A)-3'), ... | | Authors: | Dyda, F, Voth, A.R, Hickman, A.B. | | Deposit date: | 2018-06-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase.
Nucleic Acids Res., 46, 2018
|
|
6DX0
 
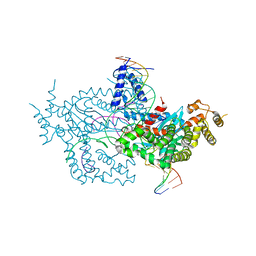 | | Hermes transposase deletion dimer complex with (A/T) DNA | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*GP*AP*GP*AP*AP*CP*AP*AP*CP*AP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*A)-3'), ... | | Authors: | Dyda, F, Voth, A.R, Hickman, A.B. | | Deposit date: | 2018-06-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase.
Nucleic Acids Res., 46, 2018
|
|
6DWY
 
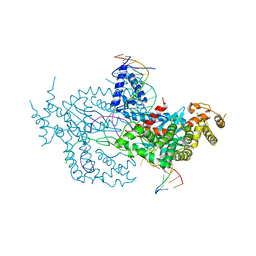 | | Hermes transposase deletion dimer complex with (C/G) DNA and Ca2+ | | Descriptor: | CALCIUM ION, DNA (26-MER), DNA (5'-D(*AP*GP*AP*GP*AP*AP*CP*AP*AP*CP*AP*AP*CP*AP*AP*G)-3'), ... | | Authors: | Dyda, F, Hickman, A.B. | | Deposit date: | 2018-06-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase.
Nucleic Acids Res., 46, 2018
|
|
1ITG
 
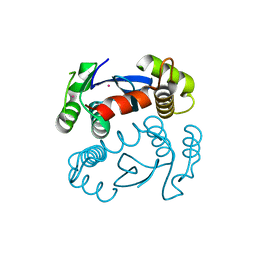 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HIV-1 INTEGRASE: SIMILARITY TO OTHER POLYNUCLEOTIDYL TRANSFERASES | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Dyda, F, Hickman, A.B, Jenkins, T.M, Engelman, A, Craigie, R, Davies, D.R. | | Deposit date: | 1994-11-21 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases.
Science, 266, 1994
|
|
8SJD
 
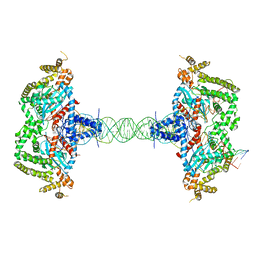 | |
6XGX
 
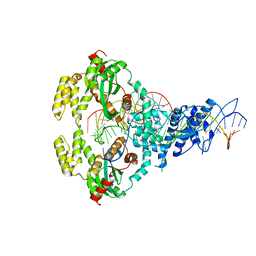 | | ISCth4 transposase, strand transfer complex 1, STC1 | | Descriptor: | DNA (21-MER), DNA (25-MER), DNA (47-MER), ... | | Authors: | Kosek, D, Dyda, F. | | Deposit date: | 2020-06-18 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of ISCth4 transpososomes reveal the role of asymmetry in copy-out/paste-in DNA transposition.
Embo J., 40, 2021
|
|
8EDG
 
 | |
7TH0
 
 | | Escherichia coli RpnA-S | | Descriptor: | Recombination-promoting nuclease RpnA | | Authors: | Zhong, A, Hickman, A.B, Storz, G, Dyda, F. | | Deposit date: | 2022-01-10 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Toxic antiphage defense proteins inhibited by intragenic antitoxin proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1AER
 
 | | DOMAIN III OF PSEUDOMONAS AERUGINOSA EXOTOXIN COMPLEXED WITH BETA-TAD | | Descriptor: | 2-(1,5-DIDEOXYRIBOSE)-4-AMIDO-THIAZOLE, ADENOSINE MONOPHOSPHATE, BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, ... | | Authors: | Li, M, Dyda, F, Benhar, I, Pastan, I, Davies, D.R. | | Deposit date: | 1995-12-11 | | Release date: | 1996-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of Pseudomonas exotoxin A complexed with a nicotinamide adenine dinucleotide analog: implications for the activation process and for ADP ribosylation
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
7MGV
 
 | | Chryseobacterium gregarium RiPP-associated ATP-grasp ligase in complex with ADP, and a leader and core peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CdnA3 Core peptide, CdnA3 Leader peptide, ... | | Authors: | Bewley, C.A, Zhao, G, Kosek, D, Dyda, F. | | Deposit date: | 2021-04-13 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural Basis for a Dual Function ATP Grasp Ligase That Installs Single and Bicyclic omega-Ester Macrocycles in a New Multicore RiPP Natural Product.
J.Am.Chem.Soc., 143, 2021
|
|
8EB5
 
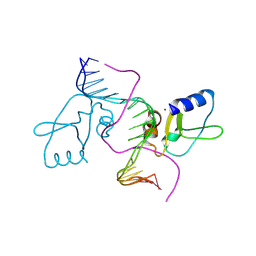 | |
1BIU
 
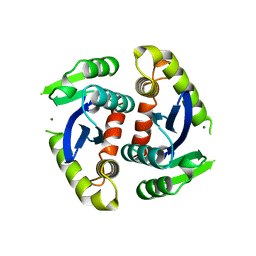 | | HIV-1 INTEGRASE CORE DOMAIN COMPLEXED WITH MG++ | | Descriptor: | HIV-1 INTEGRASE, MAGNESIUM ION | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BIZ
 
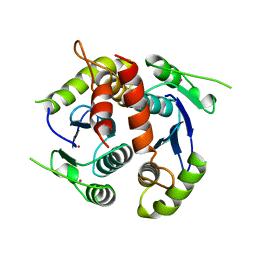 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-21 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2VJV
 
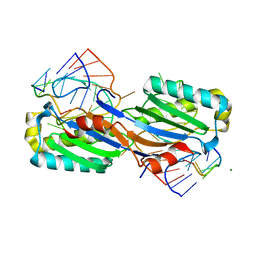 | | Crystal structure of the IS608 transposase in complex with left end 26-mer DNA hairpin and a 6-mer DNA representing the left end cleavage site | | Descriptor: | 5'-D(*DA*DA*DA*DG*DC*DC*DC*DC*DT*DA*DG*DC*DTP*DT *DT*DT*DA*DG*DC*DT*DA*DT*DG*DG*DG*DGP)-3', 5'-D(*DT*DA*DT*DT*DA*DCP)-3', MAGNESIUM ION, ... | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-12-13 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
1BIS
 
 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1PYD
 
 | |
1T0F
 
 | | Crystal Structure of the TnsA/TnsC(504-555) complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, MALONIC ACID, ... | | Authors: | Ronning, D.R, Li, Y, Perez, Z.N, Ross, P.D, Hickman, A.B, Craig, N.L, Dyda, F. | | Deposit date: | 2004-04-08 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The carboxy-terminal portion of TnsC activates the Tn7 transposase through a specific interaction with TnsA.
Embo J., 23, 2004
|
|
7LCC
 
 | | Helitron transposase bound to LTS | | Descriptor: | Helraiser K1068Q, LTS, ZINC ION | | Authors: | Kosek, D, Dyda, F. | | Deposit date: | 2021-01-10 | | Release date: | 2021-08-25 | | Last modified: | 2021-11-03 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | The large bat Helitron DNA transposase forms a compact monomeric assembly that buries and protects its covalently bound 5'-transposon end.
Mol.Cell, 81, 2021
|
|
1AIH
 
 | | CATALYTIC DOMAIN OF BACTERIOPHAGE HP1 INTEGRASE | | Descriptor: | HP1 INTEGRASE, MAGNESIUM ION, SULFATE ION | | Authors: | Hickman, A.B, Waninger, S, Scocca, J.J, Dyda, F. | | Deposit date: | 1997-04-17 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular organization in site-specific recombination: the catalytic domain of bacteriophage HP1 integrase at 2.7 A resolution.
Cell(Cambridge,Mass.), 89, 1997
|
|
