1FDN
 
 | | REFINED CRYSTAL STRUCTURE OF THE 2[4FE-4S] FERREDOXIN FROM CLOSTRIDIUM ACIDURICI AT 1.84 ANGSTROMS RESOLUTION | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Duee, E, Fanchon, E, Vicat, J, Sieker, L.C, Meyer, J, Moulis, J-M. | | Deposit date: | 1994-03-31 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Refined crystal structure of the 2[4Fe-4S] ferredoxin from Clostridium acidurici at 1.84 A resolution.
J.Mol.Biol., 243, 1994
|
|
1GAD
 
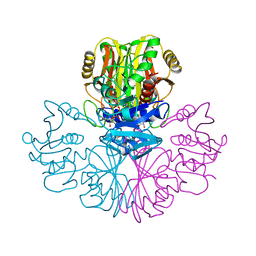 | | COMPARISON OF THE STRUCTURES OF WILD TYPE AND A N313T MUTANT OF ESCHERICHIA COLI GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASES: IMPLICATION FOR NAD BINDING AND COOPERATIVITY | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Duee, E, Olivier-Deyris, L, Fanchon, E, Corbier, C, Branlant, G, Dideberg, O. | | Deposit date: | 1995-10-24 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity.
J.Mol.Biol., 257, 1996
|
|
1GAE
 
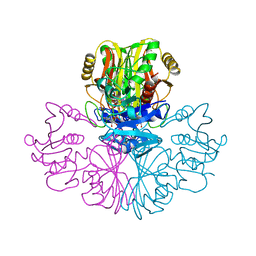 | | COMPARISON OF THE STRUCTURES OF WILD TYPE AND A N313T MUTANT OF ESCHERICHIA COLI GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASES: IMPLICATION FOR NAD BINDING AND COOPERATIVITY | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Duee, E, Olivier-Deyris, L, Fanchon, E, Corbier, C, Branlant, G, Dideberg, O. | | Deposit date: | 1995-10-24 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity.
J.Mol.Biol., 257, 1996
|
|
1BVT
 
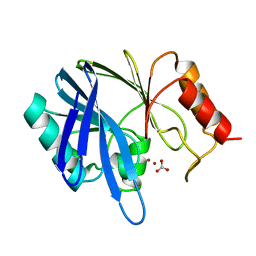 | | METALLO-BETA-LACTAMASE FROM BACILLUS CEREUS 569/H/9 | | Descriptor: | BICARBONATE ION, PROTEIN (BETA-LACTAMASE), ZINC ION | | Authors: | Carfi, A, Duee, E, Dideberg, O. | | Deposit date: | 1998-09-18 | | Release date: | 1998-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 A resolution structure of the zinc (II) beta-lactamase from Bacillus cereus.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1BMC
 
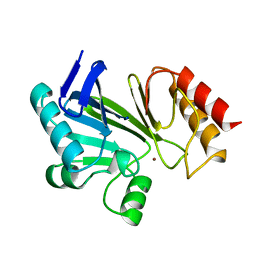 | | STRUCTURE OF A ZINC METALLO-BETA-LACTAMASE FROM BACILLUS CEREUS | | Descriptor: | METALLO-BETA-LACTAMASE, ZINC ION | | Authors: | Carfi, A, Pares, S, Duee, E, Dideberg, O. | | Deposit date: | 1995-06-16 | | Release date: | 1996-08-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold.
EMBO J., 14, 1995
|
|
2BMI
 
 | | METALLO-BETA-LACTAMASE | | Descriptor: | PROTEIN (CLASS B BETA-LACTAMASE), SODIUM ION, ZINC ION | | Authors: | Carfi, A, Duee, E, Dideberg, O. | | Deposit date: | 1998-09-17 | | Release date: | 1998-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of the ZnII beta-lactamase from Bacteroides fragilis in an orthorhombic crystal form.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1DXK
 
 | |
1QME
 
 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) | | Descriptor: | PENICILLIN-BINDING PROTEIN 2X, SULFATE ION | | Authors: | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Penicillin-Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance.
J.Mol.Biol., 299, 2000
|
|
1QMF
 
 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) ACYL-ENZYME COMPLEX | | Descriptor: | 2-[CARBOXY-(2-FURAN-2-YL-2-METHOXYIMINO-ACETYLAMINO)-METHYL]-5-METHYL-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, CEFUROXIME (OCT-3-ENE FORM), PENICILLIN-BINDING PROTEIN 2X | | Authors: | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-25 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the Penicillin Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance
J.Mol.Biol., 299, 2000
|
|
1DXH
 
 | | Catabolic ornithine carbamoyltransferase from Pseudomonas aeruginosa | | Descriptor: | ORNITHINE CARBAMOYLTRANSFERASE, SULFATE ION | | Authors: | Sainz, G, Vicat, J, Kahn, R, Duee, E, Tricot, C, Stalon, V, Dideberg, O. | | Deposit date: | 2000-01-05 | | Release date: | 2001-01-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Allosteric Active Form of Catabolic Ornithine Carbamoyltransferase from Pseudomonas Aeruginosa
To be Published
|
|
