3BKY
 
 | | Crystal Structure of Chimeric Antibody C2H7 Fab in complex with a CD20 Peptide | | Descriptor: | B-lymphocyte antigen CD20, the Fab fragment of chimeric 2H7, heavy chain, ... | | Authors: | Du, J, Zhong, C, Ding, J. | | Deposit date: | 2007-12-07 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of chimeric antibody C2H7 Fab in complex with a CD20 peptide
Mol.Immunol., 45, 2008
|
|
5WXV
 
 | |
8D8R
 
 | | SARS-CoV-2 Spike RBD in complex with DMAb 2196 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2196 heavy chain, 2196 light chain, ... | | Authors: | Du, J, Cui, J, Pallesen, J. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | DNA-delivered antibody cocktail exhibits improved pharmacokinetics and confers prophylactic protection against SARS-CoV-2.
Nat Commun, 13, 2022
|
|
8D8Q
 
 | | SARS-CoV-2 Spike RBD in complex with DMAbs 2130 and 2196 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2130 heavy chain, 2130 light chain, ... | | Authors: | Du, J, Cui, J, Pallesen, J. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | DNA-delivered antibody cocktail exhibits improved pharmacokinetics and confers prophylactic protection against SARS-CoV-2.
Nat Commun, 13, 2022
|
|
3IU3
 
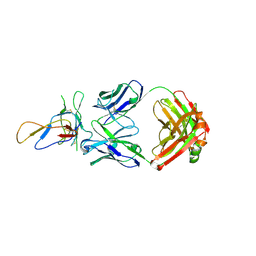 | | Crystal structure of the Fab fragment of therapeutic antibody Basiliximab in complex with IL-2Ra (CD25) ectodomain | | Descriptor: | Heavy chain of Fab fragment of Basiliximab, Interleukin-2 receptor alpha chain, Light chain of Fab fragment of Basiliximab, ... | | Authors: | Du, J, Yang, H, Wang, J, Ding, J. | | Deposit date: | 2009-08-29 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the blockage of IL-2 signaling by therapeutic antibody basiliximab
J.Immunol., 184, 2010
|
|
7N29
 
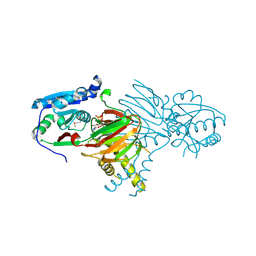 | | Structure of NAD kinase | | Descriptor: | NAD kinase 2, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Du, J, Estrella, M.A, Jeffrey, P.D, Korennykh, A.V. | | Deposit date: | 2021-05-28 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human NADK2 reveals atypical assembly and regulation of NAD kinases from animal mitochondria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3CXD
 
 | | Crystal structure of anti-osteopontin antibody 23C3 in complex with its epitope peptide | | Descriptor: | Fab fragment of anti-osteopontin antibody 23C3, Heavy chain, Light chain, ... | | Authors: | Du, J, Yang, H, Zhong, C, Ding, J. | | Deposit date: | 2008-04-24 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of recognition of human osteopontin by 23C3, a potential therapeutic antibody for treatment of rheumatoid arthritis
J.Mol.Biol., 382, 2008
|
|
3DSF
 
 | | Crystal structure of anti-osteopontin antibody 23C3 in complex with W43A mutated epitope peptide | | Descriptor: | Fab fragment of anti-osteopontin antibody 23C3, Heavy chain, Light chain, ... | | Authors: | Du, J, Zhong, C, Yang, H, Ding, J. | | Deposit date: | 2008-07-12 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of recognition of human osteopontin by 23C3, a potential therapeutic antibody for treatment of rheumatoid arthritis
J.Mol.Biol., 382, 2008
|
|
3GIZ
 
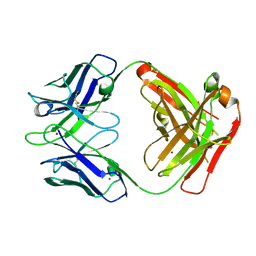 | | Crystal structure of the Fab fragment of anti-CD20 antibody Ofatumumab | | Descriptor: | Fab fragment of anti-CD20 antibody Ofatumumab, heavy chain, light chain, ... | | Authors: | Du, J, Yang, H, Ding, J. | | Deposit date: | 2009-03-07 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Fab fragment of therapeutic antibody Ofatumumab provides insights into the recognition mechanism with CD20
Mol.Immunol., 46, 2009
|
|
6PXD
 
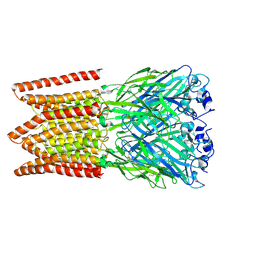 | | CryoEM structure of zebra fish alpha-1 glycine receptor, Apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glycine receptor subunit alphaZ1 | | Authors: | Du, J, Lu, W, Gouaux, E. | | Deposit date: | 2019-07-25 | | Release date: | 2021-02-10 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
4ONJ
 
 | | Crystal structure of the catalytic domain of ntDRM | | Descriptor: | DNA methyltransferase, SINEFUNGIN | | Authors: | Du, J, Patel, D.J. | | Deposit date: | 2014-01-28 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Molecular Mechanism of Action of Plant DRM De Novo DNA Methyltransferases.
Cell(Cambridge,Mass.), 157, 2014
|
|
4ONQ
 
 | |
6PUS
 
 | | Human TRPM2 bound to ADPR and calcium | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CALCIUM ION, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
6PUR
 
 | | Human TRPM2 bound to ADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
6PUU
 
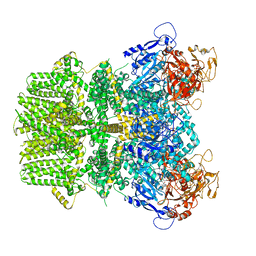 | | Human TRPM2 bound to 8-Br-cADPR and calcium | | Descriptor: | (2R,3R,4S,5R,13R,14S,15R,16R)-24-amino-18-bromo-3,4,14,15-tetrahydroxy-7,9,11,25,26-pentaoxa-17,19,22-triaza-1-azonia-8 ,10-diphosphapentacyclo[18.3.1.1^2,5^.1^13,16^.0^17,21^]hexacosa-1(24),18,20,22-tetraene-8,10-diolate 8,10-dioxide, CALCIUM ION, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
4QEN
 
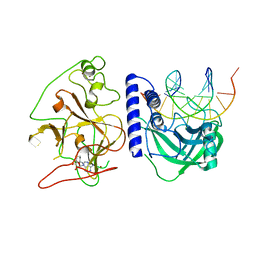 | | crystal structure of KRYPTONITE in complex with mCHH DNA and SAH | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*AP*TP*GP*AP*GP*TP*AP*CP*CP*AP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*TP*(5CM)P*AP*TP*CP*AP*GP*TP*AP*T)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Du, J, Li, S, Patel, D.J. | | Deposit date: | 2014-05-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
3JAF
 
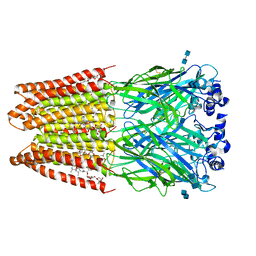 | | Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, glycine/ivermectin-bound state | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | Authors: | Du, J, Lu, W, Wu, S.P, Cheng, Y.F, Gouaux, E. | | Deposit date: | 2015-06-08 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Glycine receptor mechanism elucidated by electron cryo-microscopy.
Nature, 526, 2015
|
|
3JAE
 
 | | Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, glycine-bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | Authors: | Du, J, Lu, W, Wu, S.P, Cheng, Y.F, Gouaux, E. | | Deposit date: | 2015-06-08 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Glycine receptor mechanism elucidated by electron cryo-microscopy.
Nature, 526, 2015
|
|
4QEO
 
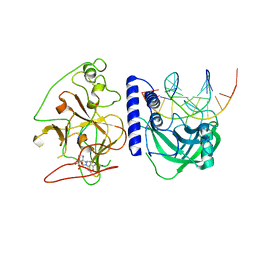 | | crystal structure of KRYPTONITE in complex with mCHH DNA, H3(1-15) peptide and SAH | | Descriptor: | DNA 5'-ACTGATGAGTACCAT-3', DNA 5'-GGTACT(5CM)ATCAGTAT-3', Histone H3, ... | | Authors: | Du, J, Li, S, Patel, D.J. | | Deposit date: | 2014-05-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
3JAD
 
 | | Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, strychnine-bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1, STRYCHNINE | | Authors: | Du, J, Lu, W, Wu, S.P, Cheng, Y.F, Gouaux, E. | | Deposit date: | 2015-06-08 | | Release date: | 2015-09-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Glycine receptor mechanism elucidated by electron cryo-microscopy.
Nature, 526, 2015
|
|
4IUU
 
 | |
4IUP
 
 | |
4IUV
 
 | |
4IUT
 
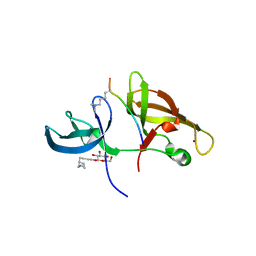 | |
4IUQ
 
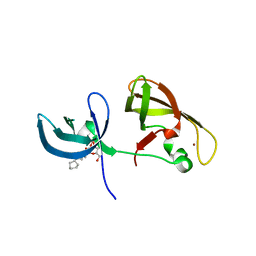 | | crystal structure of SHH1 SAWADEE domain | | Descriptor: | CYMAL-4, SAWADEE HOMEODOMAIN HOMOLOG 1, ZINC ION | | Authors: | Du, J, Patel, D.J. | | Deposit date: | 2013-01-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1.
Nature, 498, 2013
|
|
