7P9K
 
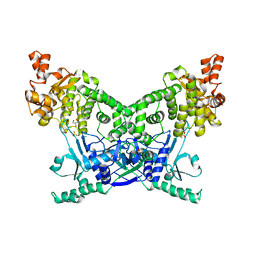 | | BrxU, GmrSD-family Type IV restriction enzyme | | Descriptor: | CHLORIDE ION, DUF262 domain-containing protein, GLYCEROL, ... | | Authors: | Picton, D.M, Luyten, Y, Morgan, R.D, Nelson, A, Smith, D.L, Dryden, D.T.F, Hinton, J.C.D, Blower, T.R. | | Deposit date: | 2021-07-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The phage defence island of a multidrug resistant plasmid uses both BREX and type IV restriction for complementary protection from viruses.
Nucleic Acids Res., 49, 2021
|
|
7P9M
 
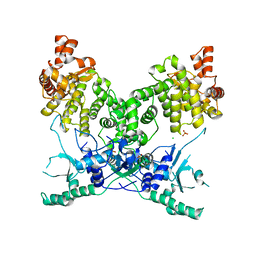 | | BrxU, GmrSD-family Type IV restriction enzyme | | Descriptor: | CHLORIDE ION, DUF262 domain-containing protein, SULFATE ION | | Authors: | Picton, D.M, Luyten, Y, Morgan, R.D, Nelson, A, Smith, D.L, Dryden, D.T.F, Hinton, J.C.D, Blower, T.R. | | Deposit date: | 2021-07-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The phage defence island of a multidrug resistant plasmid uses both BREX and type IV restriction for complementary protection from viruses.
Nucleic Acids Res., 49, 2021
|
|
2C7P
 
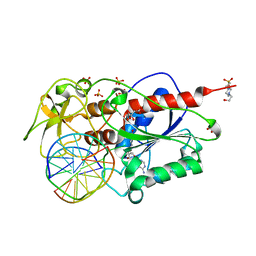 | | HhaI DNA methyltransferase complex with oligonucleotide containing 2- aminopurine opposite to the target base (GCGC:GMPC) and SAH | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-D(*G*GP*AP*TP*GP*(5CM*2PR)*CP*TP*GP*AP*C)-3', 5'-D(*G*TP*CP*AP*GP*CP*GP*CP*AP*TP*CP*C)-3', ... | | Authors: | Neely, R.K, Daujotyte, D, Grazulis, S, Magennis, S.W, Dryden, D.T.F, Klimasauskas, S, Jones, A.C. | | Deposit date: | 2005-11-25 | | Release date: | 2005-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Time-Resolved Fluorescence of 2-Aminopurine as a Probe of Base Flipping in M.HhaI-DNA Complexes.
Nucleic Acids Res., 33, 2005
|
|
2KMG
 
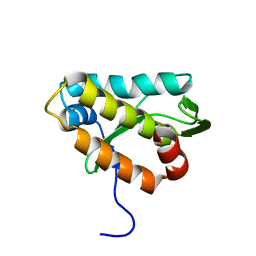 | | The structure of the KlcA and ArdB proteins show a novel fold and antirestriction activity against Type I DNA restriction systems in vivo but not in vitro | | Descriptor: | KlcA | | Authors: | Serfiotis-Mitsa, D, Herbert, A.P, Roberts, G.A, Soares, D.C, White, J.H, Blakely, G.W, Uhrin, D, Dryden, D.T.F. | | Deposit date: | 2009-07-28 | | Release date: | 2009-12-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The structure of the KlcA and ArdB proteins reveals a novel fold and antirestriction activity against Type I DNA restriction systems in vivo but not in vitro
Nucleic Acids Res., 38, 2010
|
|
2W82
 
 | | The structure of ArdA | | Descriptor: | ORF18 | | Authors: | McMahon, S.A, Roberts, G.A, Carter, L.G, Cooper, L.P, Liu, H, White, J.H, Johnson, K.A, Sanghvi, B, Oke, M, Walkinshaw, M.D, Blakely, G, Naismith, J.H, Dryden, D.T.F. | | Deposit date: | 2009-01-08 | | Release date: | 2009-01-27 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Extensive DNA Mimicry by the Arda Anti-Restriction Protein and its Role in the Spread of Antibiotic Resistance.
Nucleic Acids Res., 37, 2009
|
|
2WJ9
 
 | | ArdB | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Weikart, N.D, Roberts, G, Johnson, K.A, Oke, M, Cooper, L.P, McMahon, S.A, White, J.H, Liu, H, Carter, L.G, Walkinshaw, M.D, Blakely, G.W, Naismith, J.H, Dryden, D.T.F. | | Deposit date: | 2009-05-25 | | Release date: | 2010-08-18 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2Y7C
 
 | | Atomic model of the Ocr-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534. | | Descriptor: | GENE 0.3 PROTEIN, TYPE I RESTRICTION ENZYME ECOKI M PROTEIN, TYPE-1 RESTRICTION ENZYME ECOKI SPECIFICITY PROTEIN | | Authors: | Kennaway, C.K, Obarska-Kosinska, A, White, J.H, Tuszynska, I, Cooper, L.P, Bujnicki, J.M, Trinick, J, Dryden, D.T.F. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | The Structure of M.Ecoki Type I DNA Methyltransferase with a DNA Mimic Antirestriction Protein.
Nucleic Acids Res., 37, 2009
|
|
2Y7H
 
 | | Atomic model of the DNA-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534. | | Descriptor: | 5'-D(*GP*TP*TP*CP*AP*AP*CP*GP*TP*CP*GP*AP*CP*GP *TP*GP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*CP*AP*CP*GP*TP*CP*GP*AP*CP*GP *TP*TP*GP*AP*AP*C)-3', S-ADENOSYLMETHIONINE, ... | | Authors: | Kennaway, C.K, Obarska-Kosinska, A, White, J.H, Tuszynska, I, Cooper, L.P, Bujnicki, J.M, Trinick, J, Dryden, D.T.F. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | The Structure of M.Ecoki Type I DNA Methyltransferase with a DNA Mimic Antirestriction Protein.
Nucleic Acids Res., 37, 2009
|
|
