6OD1
 
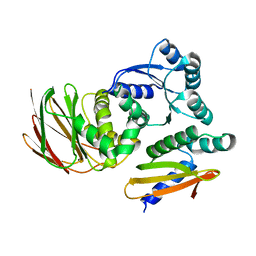 | | IraD-bound to RssB D58P variant | | Descriptor: | Anti-adapter protein IraD, Regulator of RpoS | | Authors: | Deaconescu, A.M, Dorich, V. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition of a response regulator of sigmaSstability by a ClpXP antiadaptor.
Genes Dev., 33, 2019
|
|
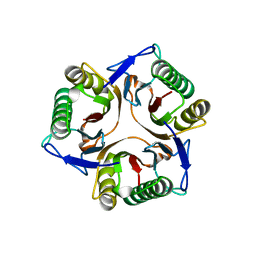
1JD1
 
 | |
4DFC
 
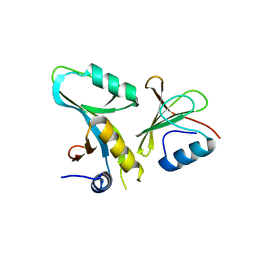 | | Core UvrA/TRCF complex | | Descriptor: | Transcription-repair-coupling factor, UvrABC system protein A | | Authors: | Deaconescu, A.M, Grigorieff, N. | | Deposit date: | 2012-01-23 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Nucleotide excision repair (NER) machinery recruitment by the transcription-repair coupling factor involves unmasking of a conserved intramolecular interface.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2EYQ
 
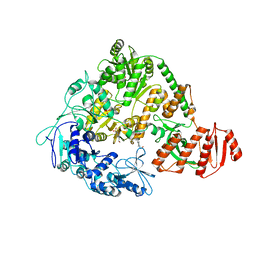 | |
7L9C
 
 | | Receiver Domain of RssB | | Descriptor: | Regulator of RpoS | | Authors: | Deaconescu, A.M, Son, J, Schwartz, J. | | Deposit date: | 2021-01-03 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Phospho-dependent signaling during the general stress response by the atypical response regulator and ClpXP adaptor RssB.
Protein Sci., 30, 2021
|
|
7LCM
 
 | | Receiver Domain of RssB bound to beryllofluoride | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Regulator of RpoS | | Authors: | Deaconescu, A.M, Schwartz, J, Son, J. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Phospho-dependent signaling during the general stress response by the atypical response regulator and ClpXP adaptor RssB.
Protein Sci., 30, 2021
|
|
8TWD
 
 | | Structure of IraM-bound RssB | | Descriptor: | ACETATE ION, Anti-adapter protein IraM, Regulator of RpoS | | Authors: | Brugger, C, Deaconescu, A.M. | | Deposit date: | 2023-08-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | IraM remodels the RssB segmented helical linker to stabilize sigma s against degradation by ClpXP.
J.Biol.Chem., 300, 2023
|
|
8T85
 
 | | Structure of RssB bound to beryllofluoride | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Brugger, C, Schwartz, J, Deaconescu, A.M. | | Deposit date: | 2023-06-21 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of phosphorylated-like RssB, the adaptor delivering sigma s to the ClpXP proteolytic machinery, reveals an interface switch for activation.
J.Biol.Chem., 299, 2023
|
|
