3BV0
 
 | |
5ZZM
 
 | |
6CJD
 
 | |
8ABY
 
 | |
8AD1
 
 | |
8ACP
 
 | |
8ABZ
 
 | |
8AC2
 
 | | RNA polymerase- post-terminated, open clamp state | | Descriptor: | DNA Non-template strand, DNA Template strand, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Dey, S, Weixlbaumer, A. | | Deposit date: | 2022-07-05 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into RNA-mediated transcription regulation in bacteria.
Mol.Cell, 82, 2022
|
|
8AC1
 
 | |
4QHT
 
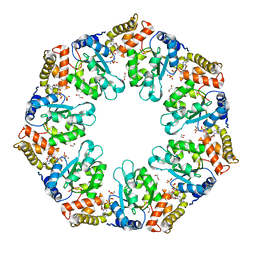 | | Crystal structure of AAA+/ sigma 54 activator domain of the flagellar regulatory protein FlrC from Vibrio cholerae in ATP analog bound state | | Descriptor: | 1,2-ETHANEDIOL, Flagellar regulatory protein C, MAGNESIUM ION, ... | | Authors: | Dey, S, Biswas, M, Sen, U, Dasgupta, J. | | Deposit date: | 2014-05-29 | | Release date: | 2014-07-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.559 Å) | | Cite: | Unique ATPase site architecture triggers cis-mediated synchronized ATP binding in heptameric AAA+-ATPase domain of flagellar regulatory protein FlrC
J.Biol.Chem., 290, 2015
|
|
3FPA
 
 | |
4QHS
 
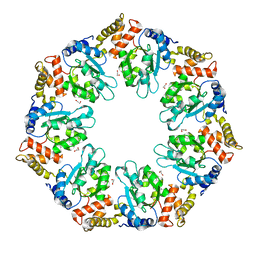 | | Crystal structure of AAA+sigma 54 activator domain of the flagellar regulatory protein FlrC of Vibrio cholerae in nucleotide free state | | Descriptor: | 1,2-ETHANEDIOL, Flagellar regulatory protein C | | Authors: | Dey, S, Biswas, M, Sen, U, Dasgupta, J. | | Deposit date: | 2014-05-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unique ATPase site architecture triggers cis-mediated synchronized ATP binding in heptameric AAA+-ATPase domain of flagellar regulatory protein FlrC
J.Biol.Chem., 290, 2015
|
|
3FMI
 
 | |
3DU4
 
 | |
1YGY
 
 | |
3DRD
 
 | |
3FMF
 
 | |
3DOD
 
 | |
3DC2
 
 | |
3FGN
 
 | |
3DDN
 
 | |
3LV2
 
 | |
2P9E
 
 | |
2P9C
 
 | |
2PA3
 
 | |
