1I3Q
 
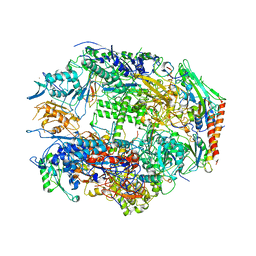 | | RNA POLYMERASE II CRYSTAL FORM I AT 3.1 A RESOLUTION | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.5KD POLYPEPTIDE, ... | | Authors: | Cramer, P, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2001-02-15 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution.
Science, 292, 2001
|
|
1I50
 
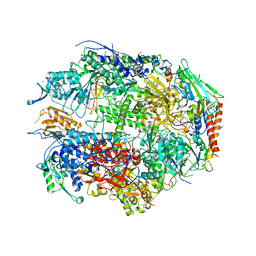 | | RNA POLYMERASE II CRYSTAL FORM II AT 2.8 A RESOLUTION | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.5KD POLYPEPTIDE, ... | | Authors: | Cramer, P, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2001-02-23 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution.
Science, 292, 2001
|
|
1A3Q
 
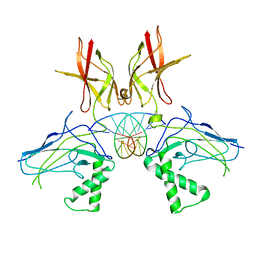 | | HUMAN NF-KAPPA-B P52 BOUND TO DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*AP*AP*TP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*AP*TP*TP*CP*CP*CP*C)-3'), PROTEIN (NUCLEAR FACTOR KAPPA-B P52) | | Authors: | Cramer, P, Larson, C.J, Verdine, G.L, Muller, C.W. | | Deposit date: | 1998-01-23 | | Release date: | 1998-06-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the human NF-kappaB p52 homodimer-DNA complex at 2.1 A resolution.
EMBO J., 16, 1997
|
|
1BRI
 
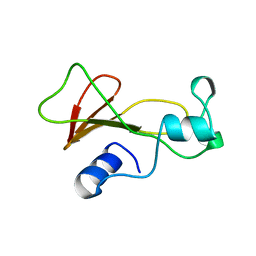 | | BARNASE MUTANT WITH ILE 76 REPLACED BY ALA | | Descriptor: | BARNASE | | Authors: | Cramer, P.C, Buckle, A, Fersht, A. | | Deposit date: | 1995-03-09 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and energetic responses to cavity-creating mutations in hydrophobic cores: observation of a buried water molecule and the hydrophilic nature of such hydrophobic cavities.
Biochemistry, 35, 1996
|
|
1BRK
 
 | | BARNASE MUTANT WITH ILE 96 REPLACED BY ALA | | Descriptor: | BARNASE, ZINC ION | | Authors: | Cramer, P.C, Buckle, A, Fersht, A. | | Deposit date: | 1995-03-09 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and energetic responses to cavity-creating mutations in hydrophobic cores: observation of a buried water molecule and the hydrophilic nature of such hydrophobic cavities.
Biochemistry, 35, 1996
|
|
1Y1W
 
 | | Complete RNA Polymerase II elongation complex | | Descriptor: | 5'-D(*AP*AP*GP*TP*AP*CP*T)-3', 5'-D(P*AP*GP*TP*AP*CP*TP*TP*AP*CP*GP*CP*CP*TP*GP*GP*TP*CP*AP*T)-3', 5'-R(*AP*AP*GP*AP*CP*CP*AP*GP*GP*C)-3', ... | | Authors: | Cramer, P, Kettenberger, H, Armache, K.-J. | | Deposit date: | 2004-11-19 | | Release date: | 2005-01-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Complete RNA Polymerase II Elongation Complex Structure and Its Interactions with NTP and TFIIS
Mol.Cell, 16, 2004
|
|
1BRJ
 
 | | BARNASE MUTANT WITH ILE 88 REPLACED BY ALA | | Descriptor: | BARNASE, ZINC ION | | Authors: | Cramer, P.C, Buckle, A, Fersht, A. | | Deposit date: | 1995-03-09 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and energetic responses to cavity-creating mutations in hydrophobic cores: observation of a buried water molecule and the hydrophilic nature of such hydrophobic cavities.
Biochemistry, 35, 1996
|
|
1BRH
 
 | | BARNASE MUTANT WITH LEU 14 REPLACED BY ALA | | Descriptor: | BARNASE, ZINC ION | | Authors: | Cramer, P.C, Buckle, A, Fersht, A. | | Deposit date: | 1995-03-09 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and energetic responses to cavity-creating mutations in hydrophobic cores: observation of a buried water molecule and the hydrophilic nature of such hydrophobic cavities.
Biochemistry, 35, 1996
|
|
1Y1Y
 
 | | RNA Polymerase II-TFIIS-DNA/RNA complex | | Descriptor: | 5'-D(P*TP*AP*CP*GP*CP*CP*T)-3', 5'-R(P*AP*GP*GP*C)-3', DNA-directed RNA polymerase II 13.6 kDa polypeptide, ... | | Authors: | Cramer, P, Kettenberger, H, Armache, K.-J. | | Deposit date: | 2004-11-19 | | Release date: | 2004-12-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS.
Mol.Cell, 16, 2004
|
|
1BVO
 
 | | DORSAL HOMOLOGUE GAMBIF1 BOUND TO DNA | | Descriptor: | DNA DUPLEX, TRANSCRIPTION FACTOR GAMBIF1 | | Authors: | Cramer, P, Varrot, A, Barillas-Mury, C, Kafatos, F.C, Mueller, C.W. | | Deposit date: | 1998-09-16 | | Release date: | 1999-07-12 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the specificity domain of the Dorsal homologue Gambif1 bound to DNA.
Structure Fold.Des., 7, 1999
|
|
4A93
 
 | | RNA Polymerase II elongation complex containing a CPD Lesion | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP*CP*T*TTP*TP*TP*CP*C BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*TP)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP*AP)-3', ... | | Authors: | Walmacq, C, Cheung, A.C.M, Kireeva, M.L, Lubkowska, L, Ye, C, Gotte, D, Strathern, J.N, Carell, T, Cramer, P, Kashlev, M. | | Deposit date: | 2011-11-23 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanism of Translesion Transcription by RNA Polymerase II and its Role in Cellular Resistance to DNA Damage.
Mol.Cell, 46, 2012
|
|
3C0T
 
 | | Structure of the Schizosaccharomyces pombe Mediator subcomplex Med8C/18 | | Descriptor: | Mediator of RNA polymerase II transcription subunit 18, Mediator of RNA polymerase II transcription subunit 8 | | Authors: | Lariviere, L, Seizl, M, van Wageningen, S, Roether, S, van de Pasch, L, Feldmann, H, Straesser, K, Hahn, S, Holstege, C.P, Cramer, P. | | Deposit date: | 2008-01-21 | | Release date: | 2008-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-system correlation identifies a gene regulatory Mediator submodule
Genes Dev., 22, 2008
|
|
4V1O
 
 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
4V1N
 
 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
4V1M
 
 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | 5'-D(*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP)-3', 5'-D(*CP*CP*AP*GP*GP*AP)-3', 5'-D(*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*TP*TP*TP*CP *CP*BRUP*GP*GP*TP*C)-3', ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
6ERQ
 
 | | Structure of the human mitochondrial transcription initiation complex at the HSP promoter | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, Dimethyladenosine transferase 2, ... | | Authors: | Hillen, H.S, Morozov, Y.I, Sarfallah, A, Temiakov, D, Cramer, P. | | Deposit date: | 2017-10-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural Basis of Mitochondrial Transcription Initiation.
Cell, 171, 2017
|
|
6ERO
 
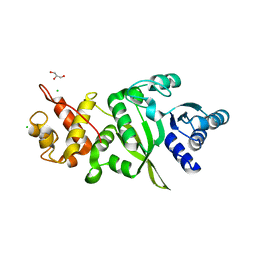 | | Structure of human TFB2M | | Descriptor: | CHLORIDE ION, Dimethyladenosine transferase 2, mitochondrial,Dimethyladenosine transferase 2, ... | | Authors: | Hillen, H.S, Morozov, Y.I, Sarfallah, A, Temiakov, D, Cramer, P. | | Deposit date: | 2017-10-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Mitochondrial Transcription Initiation.
Cell, 171, 2017
|
|
6I84
 
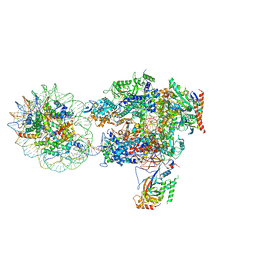 | | Structure of transcribing RNA polymerase II-nucleosome complex | | Descriptor: | DNA (158-MER), DNA (170-MER), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Farnung, L, Vos, S.M, Cramer, P. | | Deposit date: | 2018-11-19 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of transcribing RNA polymerase II-nucleosome complex.
Nat Commun, 9, 2018
|
|
9GD1
 
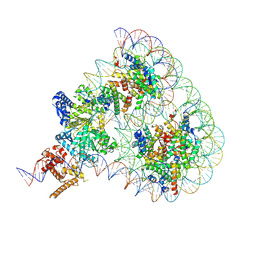 | | Structure of Chd1 bound to a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GD2
 
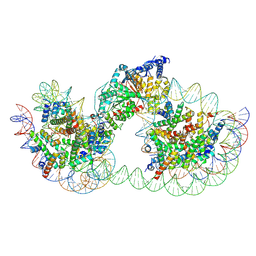 | | Structure of Chd1 bound to a dinucleosome with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GD0
 
 | | Structure of a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | DNA (250-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
4YM7
 
 | | RNA polymerase I structure with an alternative dimer hinge | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Kostrewa, D, Kuhn, C.-D, Engel, C, Cramer, P. | | Deposit date: | 2015-03-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | An alternative RNA polymerase I structure reveals a dimer hinge.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
9GD3
 
 | | Structure of a mononucleosome bound by one copy of Chd1 with the DBD on the exit-side DNA. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
5OT2
 
 | | RNA polymerase II elongation complex in the presence of 3d-Napht-A | | Descriptor: | 2-ethyl-7-methoxy-naphthalene, DNA non-template strand, DNA template strand, ... | | Authors: | Malvezzi, S, Farnung, L, Aloisi, C, Angelov, T, Cramer, P, Sturla, S.J. | | Deposit date: | 2017-08-19 | | Release date: | 2017-11-01 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of RNA polymerase II stalling by DNA alkylation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5OQM
 
 | | STRUCTURE OF YEAST TRANSCRIPTION PRE-INITIATION COMPLEX WITH TFIIH AND CORE MEDIATOR | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Schilbach, S, Hantsche, M, Tegunov, D, Dienemann, C, Wigge, C, Urlaub, H, Cramer, P. | | Deposit date: | 2017-08-13 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structures of transcription pre-initiation complex with TFIIH and Mediator.
Nature, 551, 2017
|
|
