1TI3
 
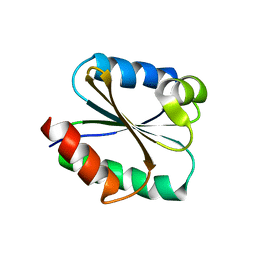 | | Solution structure of the Thioredoxin h1 from poplar, a CPPC active site variant | | Descriptor: | thioredoxin H | | Authors: | Coudevylle, N, Thureau, A, Hemmerlin, C, Gelhaye, E, Jacquot, J.P, Cung, M.T. | | Deposit date: | 2004-06-02 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a natural CPPC active site variant, the reduced form of thioredoxin h1 from poplar.
Biochemistry, 44, 2005
|
|
2KT4
 
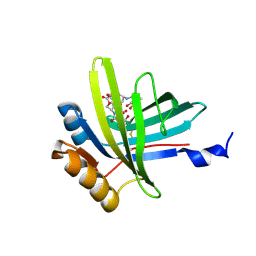 | | Lipocalin Q83 is a Siderocalin | | Descriptor: | Extracellular fatty acid-binding protein, GALLIUM (III) ION, N,N',N''-[(3S,7S,11S)-2,6,10-trioxo-1,5,9-trioxacyclododecane-3,7,11-triyl]tris(2,3-dihydroxybenzamide) | | Authors: | Coudevylle, N, Geist, L, Hartl, M, Kontaxis, G, Bister, K, Konrat, R. | | Deposit date: | 2010-01-18 | | Release date: | 2010-09-08 | | Last modified: | 2016-01-27 | | Method: | SOLUTION NMR | | Cite: | The v-myc-induced Q83 lipocalin is a siderocalin.
J.Biol.Chem., 285, 2010
|
|
2LBV
 
 | | Siderocalin Q83 reveals a dual ligand binding mode | | Descriptor: | ARACHIDONIC ACID, Extracellular fatty acid-binding protein, GALLIUM (III) ION, ... | | Authors: | Coudevylle, N, Hoetzinger, M, Geist, L, Kontaxis, G, Bister, K, Konrat, R. | | Deposit date: | 2011-04-07 | | Release date: | 2012-02-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Lipocalin Q83 reveals a dual ligand binding mode with potential implications for the functions of siderocalins
Biochemistry, 50, 2011
|
|
2GT3
 
 | |
2IEM
 
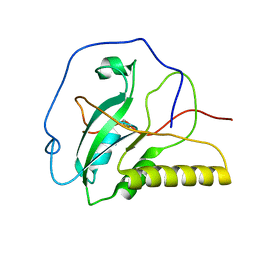 | | Solution structure of an oxidized form (Cys51-Cys198) of E. coli Methionine Sulfoxide Reductase A (MsrA) | | Descriptor: | Peptide methionine sulfoxide reductase msrA | | Authors: | Coudevylle, N, Antoine, M, Bouguet-Bonnet, S, Mutzenhardt, P, Boschi-Muller, S, Branlant, G, Cung, M.T. | | Deposit date: | 2006-09-19 | | Release date: | 2007-02-13 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Reduced Form and an Oxidized Form of E. coli Methionine Sulfoxide Reductase A (MsrA): Structural Insight of the MsrA Catalytic Cycle.
J.Mol.Biol., 366, 2007
|
|
2K3H
 
 | | Structural determinants for Ca2+ and PIP2 binding by the C2A domain of rabphilin-3A | | Descriptor: | CALCIUM ION, Rabphilin-3A | | Authors: | Coudevylle, N, Montaville, P, Leonov, A, Zweckstetter, M, Becker, S. | | Deposit date: | 2008-05-08 | | Release date: | 2008-10-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Determinants for Ca2+ and Phosphatidylinositol 4,5-Bisphosphate Binding by the C2A Domain of Rabphilin-3A.
J.Biol.Chem., 283, 2008
|
|
2KDU
 
 | | Structural basis of the Munc13-1/Ca2+-Calmodulin interaction: A novel 1-26 calmodulin binding motif with a bipartite binding mode | | Descriptor: | CALCIUM ION, Calmodulin, Protein unc-13 homolog A | | Authors: | Rodriguez-Castaneda, F.A, Maestre-Martinez, M, Coudevylle, N, Dimova, K, Jahn, O, Junge, H, Becker, S, Brose, N, Carlomagno, T, Griesinger, C. | | Deposit date: | 2009-01-19 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Modular architecture of Munc13/calmodulin complexes: dual regulation by Ca2+ and possible function in short-term synaptic plasticity.
Embo J., 29, 2010
|
|
