3GZC
 
 | | Structure of human selenocysteine lyase | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Selenocysteine lyase | | Authors: | Collins, R, Hogbom, M, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Karlberg, T, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Uppenberg, J, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-07 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical discrimination between selenium and sulfur 1: a single residue provides selenium specificity to human selenocysteine lyase.
Plos One, 7, 2012
|
|
3COG
 
 | | Crystal structure of human cystathionase (Cystathionine gamma lyase) in complex with DL-propargylglycine | | Descriptor: | (2S)-2-aminopent-4-enoic acid, Cystathionine gamma-lyase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Collins, R, Karlberg, T, Lehtio, L, Arrowsmith, C.H, Berglund, H, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Schuler, H, Svensson, L, Thorsell, A.G, Tresaugues, L, Van den Berg, S, Sagermark, J, Busam, R.D, Welin, M, Weigelt, J, Wikstrom, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-28 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the inhibition mechanism of human cystathionine gamma-lyase, an enzyme responsible for the production of H(2)S.
J.Biol.Chem., 284, 2009
|
|
2JIS
 
 | | Human cysteine sulfinic acid decarboxylase (CSAD) in complex with PLP. | | Descriptor: | CYSTEINE SULFINIC ACID DECARBOXYLASE, NITRATE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Collins, R, Moche, M, Arrowsmith, C, Berglund, H, Busam, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-30 | | Release date: | 2007-08-28 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Human Cysteine Sulfinic Acid Decarboxylase (Csad)
To be Published
|
|
3B7B
 
 | | EuHMT1 (Glp) Ankyrin Repeat Domain (Structure 1) | | Descriptor: | Euchromatic histone-lysine N-methyltransferase 1, SULFATE ION | | Authors: | Collins, R.E, Horton, J.R, Cheng, X. | | Deposit date: | 2007-10-30 | | Release date: | 2008-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules
Nat.Struct.Mol.Biol., 15, 2008
|
|
3B95
 
 | | EuHMT1 (Glp) Ankyrin Repeat Domain (Structure 2) | | Descriptor: | Euchromatic histone-lysine N-methyltransferase 1, Histone H3 N-terminal Peptide, SULFATE ION | | Authors: | Collins, R.E, Horton, J.R, Cheng, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules.
Nat.Struct.Mol.Biol., 15, 2008
|
|
6VOW
 
 | |
6VOX
 
 | |
6Q81
 
 | | Structure of P-glycoprotein(ABCB1) in the post-hydrolytic state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, P-glycoprotein (ABCB1) | | Authors: | Ford, R.C, Thonghin, N, Collins, R.F, Barbieri, A, Shafi, T, Siebert, A. | | Deposit date: | 2018-12-13 | | Release date: | 2018-12-26 | | Last modified: | 2019-04-03 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Novel features in the structure of P-glycoprotein (ABCB1) in the post-hydrolytic state as determined at 7.9 angstrom resolution.
Bmc Struct.Biol., 18, 2018
|
|
6XIZ
 
 | | Crystal structure of multi-copper oxidase from Pediococcus acidilactici | | Descriptor: | BENZAMIDINE, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Pardo, I, Soares, A.S, Collins, R, Partowmah, S.H, Coler, E.A. | | Deposit date: | 2020-06-22 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis and biochemical properties of laccase enzymes from two Pediococcus species.
Microb Biotechnol, 14, 2021
|
|
6XJ0
 
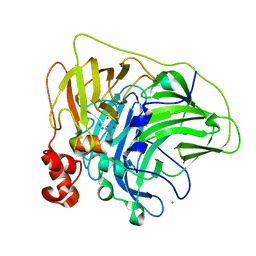 | | Crystal structure of multi-copper oxidase from Pediococcus pentosaceus | | Descriptor: | CHLORIDE ION, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Pardo, I, Soares, A.S, Collins, R, Partowmah, S.H, Coler, E.A. | | Deposit date: | 2020-06-22 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural analysis and biochemical properties of laccase enzymes from two Pediococcus species.
Microb Biotechnol, 14, 2021
|
|
7OTG
 
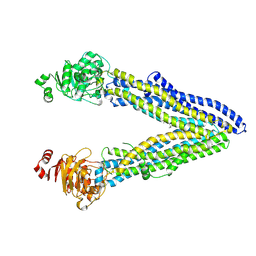 | | Structure of ABCB1/P-glycoprotein in the presence of the CFTR potentiator ivacaftor | | Descriptor: | Multidrug resistance protein 1A, N-(2,4-di-tert-butyl-5-hydroxyphenyl)-4-oxo-1,4-dihydroquinoline-3-carboxamide | | Authors: | Ford, R.C, Barbieri, A, Thonghin, N, Shafi, T, Prince, S.M, Collins, R.F. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structure of ABCB1/P-Glycoprotein in the Presence of the CFTR Potentiator Ivacaftor.
Membranes (Basel), 11, 2021
|
|
7OTI
 
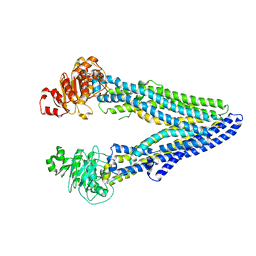 | | Structure of ABCB1/P-glycoprotein in apo state | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ford, R.C, Barbieri, A, Thonghin, N, Shafi, T, Prince, S.M, Collins, R.F. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of ABCB1/P-Glycoprotein in the Presence of the CFTR Potentiator Ivacaftor.
Membranes (Basel), 11, 2021
|
|
4ZDW
 
 | |
4Z8Y
 
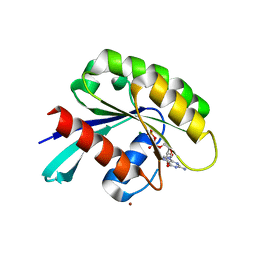 | | Crystal structure of Rab GTPase Sec4p mutant - S29V | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein SEC4, ... | | Authors: | Rinadi, F.C, Collins, R.N. | | Deposit date: | 2015-04-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New insights into the molecular mechanism of the Rab GTPase Sec4p activation.
Bmc Struct.Biol., 15, 2015
|
|
5NBZ
 
 | | Wzz dodecamer fitted by MDFF to the Wzz experimental map from cryo-EM | | Descriptor: | WzzB | | Authors: | Ford, R.C, Kargas, V, Collins, R.F, Whitfield, C, Clarke, B.R, Siebert, A, Bond, P.J, Clare, D.K. | | Deposit date: | 2017-03-02 | | Release date: | 2017-04-05 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Full-length, Oligomeric Structure of Wzz Determined by Cryoelectron Microscopy Reveals Insights into Membrane-Bound States.
Structure, 25, 2017
|
|
5N6W
 
 | | Retinoschisin R141H Mutant | | Descriptor: | Retinoschisin | | Authors: | Ramsay, E.P, Collins, R.F, Owens, T.W, Siebert, C.A, Jones, R.P.O, Roseman, A, Wang, T, Baldock, C. | | Deposit date: | 2017-02-16 | | Release date: | 2017-04-12 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of X-linked retinoschisis mutations reveals distinct classes which differentially effect retinoschisin function
Human Molecular Genetics, 25, 2016
|
|
4IRU
 
 | | Crystal Structure of lepB GAP core in a transition state mimetic complex with Rab1A and ALF3 | | Descriptor: | ACETATE ION, ALUMINUM FLUORIDE, GLYCEROL, ... | | Authors: | Mishra, A.K, Delcampo, C.M, Collins, R.E, Roy, C.R, Lambright, D.G. | | Deposit date: | 2013-01-15 | | Release date: | 2013-07-10 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Legionella pneumophila GTPase Activating Protein LepB Accelerates Rab1 Deactivation by a Non-canonical Hydrolytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
6EJF
 
 | |
6F8L
 
 | |
1PZD
 
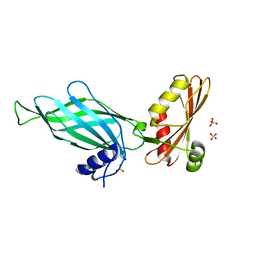 | | Structural Identification of a conserved appendage domain in the carboxyl-terminus of the COPI gamma-subunit. | | Descriptor: | Coatomer gamma subunit, SULFATE ION | | Authors: | Hoffman, G.R, Rahl, P.B, Collins, R.N, Cerione, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Conserved Structural Motifs in Intracellular Trafficking Pathways. Structure of the gammaCOP Appendage Domain.
Mol.Cell, 12, 2003
|
|
1UWX
 
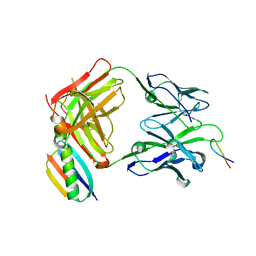 | | P1.2 serosubtype antigen derived from N. meningitidis PorA in complex with Fab fragment | | Descriptor: | ANTIBODY, CLASS 1 OUTER MEMBRANE PROTEIN VARIABLE REGION 2, PROTEIN G-PRIME | | Authors: | Tzitzilonis, C, Prince, S.M, Collins, R.F, Maiden, M.C.J, Feavers, I.M, Derrick, J.P. | | Deposit date: | 2004-02-12 | | Release date: | 2005-06-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Variation and Immune Recognition of the P1.2 Subtype Meningococcal Antigen.
Proteins: Struct., Funct., Bioinf., 62, 2005
|
|
6GDI
 
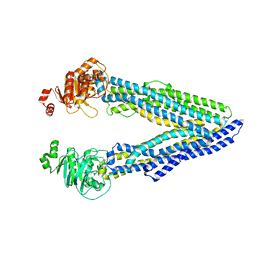 | | Structure of P-glycoprotein(ABCB1) in the post-hydrolytic state | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ford, R.C, Thonghin, N, Collins, R.F, Barbieri, A, Shafi, T, Siebert, A. | | Deposit date: | 2018-04-23 | | Release date: | 2018-05-23 | | Last modified: | 2019-04-03 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Novel features in the structure of P-glycoprotein (ABCB1) in the post-hydrolytic state as determined at 7.9 angstrom resolution.
Bmc Struct.Biol., 18, 2018
|
|
1U2Z
 
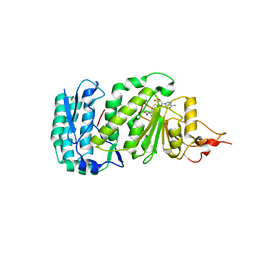 | | Crystal structure of histone K79 methyltransferase Dot1p from yeast | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sawada, K, Yang, Z, Horton, J.R, Collins, R.E, Zhang, X, Cheng, X. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase
J.Biol.Chem., 279, 2004
|
|
1OW9
 
 | | NMR Structure of the Active Conformation of the VS Ribozyme Cleavage Site | | Descriptor: | A mimic of the VS Ribozyme Hairpin Substrate | | Authors: | Hoffmann, B, Mitchell, G.T, Gendron, P, Major, F, Andersen, A.A, Collins, R.A, Legault, P. | | Deposit date: | 2003-03-28 | | Release date: | 2003-05-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Active Conformation of the Varkud satellite Ribozyme Cleavage Site
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3GZD
 
 | | Human selenocysteine lyase, P1 crystal form | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, NITRATE ION, Selenocysteine lyase | | Authors: | Karlberg, T, Hogbom, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-07 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical discrimination between selenium and sulfur 1: a single residue provides selenium specificity to human selenocysteine lyase.
Plos One, 7, 2012
|
|
