7JY2
 
 | | Z-DNA joint X-ray/Neutron | | Descriptor: | Chains: A,B | | Authors: | Harp, J.M, Coates, L, Egli, M. | | Deposit date: | 2020-08-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | Cite: | Water structure around a left-handed Z-DNA fragment analyzed by cryo neutron crystallography.
Nucleic Acids Res., 49, 2021
|
|
6V8Y
 
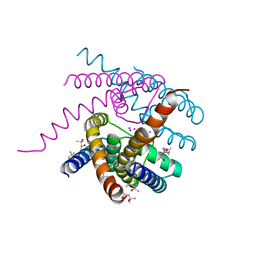 | | Structure of a Sodium Potassium ion Channel | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Roy, R, Weiss, K.L, Coates, L. | | Deposit date: | 2019-12-12 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural plasticity of the selectivity filter in a nonselective ion channel.
Iucrj, 8, 2021
|
|
6EYM
 
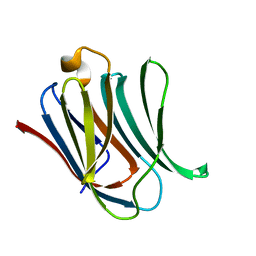 | | Neutron crystal structure of perdeuterated galectin-3C in complex with lactose | | Descriptor: | Galectin-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Manzoni, F, Coates, L, Blakeley, M.P, Oksanen, E, Logan, D.T. | | Deposit date: | 2017-11-13 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Elucidation of Hydrogen Bonding Patterns in Ligand-Free, Lactose- and Glycerol-Bound Galectin-3C by Neutron Crystallography to Guide Drug Design.
J. Med. Chem., 61, 2018
|
|
7KKU
 
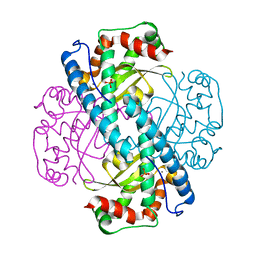 | | X-ray Counterpart to Neutron Structure of Oxidized Human MnSOD | | Descriptor: | MANGANESE (III) ION, PHOSPHATE ION, Superoxide dismutase [Mn], ... | | Authors: | Azadmanesh, J, Lutz, W.E, Coates, L, Weiss, K.L, Borgstahl, G.E.O. | | Deposit date: | 2020-10-28 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Direct detection of coupled proton and electron transfers in human manganese superoxide dismutase.
Nat Commun, 12, 2021
|
|
7KKW
 
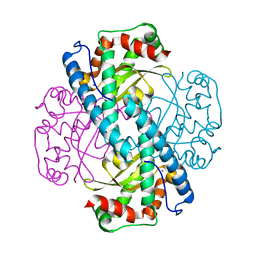 | | Neutron structure of Reduced Human MnSOD | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial, ... | | Authors: | Azadmanesh, J, Lutz, W.E, Coates, L, Weiss, K.L, Borgstahl, G.E.O. | | Deposit date: | 2020-10-28 | | Release date: | 2021-04-21 | | Last modified: | 2024-04-10 | | Method: | NEUTRON DIFFRACTION (2.3 Å) | | Cite: | Direct detection of coupled proton and electron transfers in human manganese superoxide dismutase.
Nat Commun, 12, 2021
|
|
7KKS
 
 | | Neutron structure of Oxidized Human MnSOD | | Descriptor: | MANGANESE (III) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Azadmanesh, J, Lutz, W.E, Coates, L, Weiss, K.L, Borgstahl, G.E.O. | | Deposit date: | 2020-10-28 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Direct detection of coupled proton and electron transfers in human manganese superoxide dismutase.
Nat Commun, 12, 2021
|
|
6C78
 
 | | Substrate Binding Induces Conformational Changes In A Class A Beta Lactamase That Primes It For Catalysis | | Descriptor: | Beta-lactamase Toho-1 | | Authors: | Langan, P.S, Vandavasi, V.G, Cooper, S.J, Weiss, K.L, Ginell, S.L, Parks, J.M, Coates, L. | | Deposit date: | 2018-01-22 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Substrate Binding Induces Conformational Changes in a Class A Beta-lactamase That Prime It for Catalysis
Acs Catalysis, 8, 2018
|
|
6DZ1
 
 | | Studies of Ion Transport in K+ Channels | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein | | Authors: | Langan, P.S, Vandavasi, V.G, Weiss, K.L, Wagner, A, Duman, R, El Omari, K, Afonine, P.V, Coates, L. | | Deposit date: | 2018-07-02 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Anomalous X-ray diffraction studies of ion transport in K+channels.
Nat Commun, 9, 2018
|
|
8GFN
 
 | | Room temperature X-ray structure of truncated SARS-CoV-2 main protease C145A mutant, residues 1-304, in complex with BBH1 | | Descriptor: | (1R,2S,5S)-N-{(2S)-1-(1,3-benzothiazol-2-yl)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Kovalevsky, A, Coates, L. | | Deposit date: | 2023-03-08 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the catalytic dyad of SARS-CoV-2 main protease to binding covalent and noncovalent inhibitors.
J.Biol.Chem., 299, 2023
|
|
8GFO
 
 | |
8GFR
 
 | | Room temperature X-ray structure of truncated SARS-CoV-2 main protease C145A mutant, residues 1-304, in complex with NBH2 | | Descriptor: | (1R,2S,5S)-N-{(1S)-1-cyano-2-[(3S)-2-oxopyrrolidin-3-yl]ethyl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Kovalevsky, A, Coates, L. | | Deposit date: | 2023-03-08 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of the catalytic dyad of SARS-CoV-2 main protease to binding covalent and noncovalent inhibitors.
J.Biol.Chem., 299, 2023
|
|
6XKF
 
 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with oxidized Cys145 (Sulfenic acid cysteine). | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Coates, L, Kovalevskyi, A.Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with oxidized Cys145 (Sulfenic acid cysteine).
To Be Published
|
|
6XKH
 
 | | THE 1.28A CRYSTAL STRUCTURE OF 3CL MAINPRO OF SARS-COV-2 WITH OXIDIZED C145 (sulfinic acid cysteine) | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, ACETATE ION, ... | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Coates, L, Kovalevsky, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | THE 1.28A CRYSTAL STRUCTURE OF 3CL MAINPRO OF SARS-COV-2 WITH OXIDIZED C145 (sulfinic acid cysteine)
To Be Published
|
|
5KSC
 
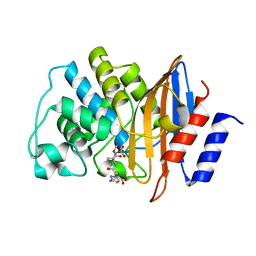 | | E166A/R274N/R276N Toho-1 Beta-lactamase aztreonam acyl-enzyme intermediate | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, Beta-lactamase Toho-1 | | Authors: | Vandavasi, V.G, Langan, P.S, Weiss, K, Parks, J.M, Cooper, J.B, Ginell, S.L, Coates, L. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2019-12-04 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | Active-Site Protonation States in an Acyl-Enzyme Intermediate of a Class A beta-Lactamase with a Monobactam Substrate.
Antimicrob. Agents Chemother., 61, 2017
|
|
8GFU
 
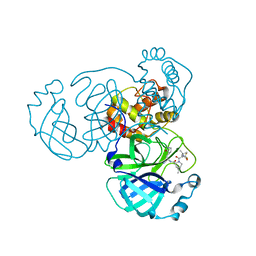 | |
7SI9
 
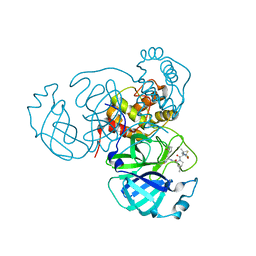 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with PF-07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-10-12 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TEH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TFR
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with NBH-2 | | Descriptor: | (1R,2S,5S)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TDU
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-1 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxo(1-~2~H)pyrrolidin-3-yl]propan-2-yl}-3-{N-[tert-butyl(~2~H)carbamoyl]-3-methyl-L-(N-~2~H)valyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-(~2~H)carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
6WQF
 
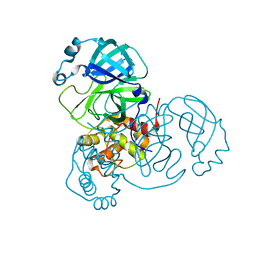 | |
6XQT
 
 | | Room-temperature X-ray Crystal structure of SARS-CoV-2 main protease in complex with Narlaprevir | | Descriptor: | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropylamino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kneller, D.W, Kovalevsky, A, Coates, L. | | Deposit date: | 2020-07-10 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Malleability of the SARS-CoV-2 3CL M pro Active-Site Cavity Facilitates Binding of Clinical Antivirals.
Structure, 28, 2020
|
|
5G18
 
 | | Direct Observation of Active-site Protonation States in a Class A beta lactamase with a monobactam substrate | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, BETA-LACTAMASE CTX-M-97, SULFATE ION | | Authors: | Vandavasi, V.G, Weiss, K.L, Parks, J.M, Cooper, J.B, Ginell, S.L, Coates, L. | | Deposit date: | 2016-03-23 | | Release date: | 2016-11-09 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Active-Site Protonation States in an Acyl-Enzyme Intermediate of a Class A beta-Lactamase with a Monobactam Substrate.
Antimicrob. Agents Chemother., 61, 2017
|
|
6XQU
 
 | |
5TY5
 
 | | Neutron structure from microgravity-grown crystals of Inorganic Pyrophosphatase from Thermococcus theoreducens | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Inoguchi, N, Coates, L, Morris, M.L, Singhal, A, Monaco, D.A, Garcia-Ruiz, J.M, Pusey, M.L, Ng, J.D. | | Deposit date: | 2016-11-18 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (2.3 Å) | | Cite: | Structure-function analysis of the neutron crystallographic structure of inorganic pyrophosphatase determined from microgravity-grown crystals
Acta Crystallogr.,Sect.A, 73, 2017
|
|
7UKK
 
 | |
