1OKO
 
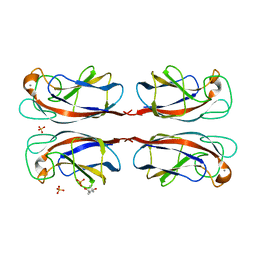 | | Crystal structure of Pseudomonas Aeruginosa Lectin 1 complexed with galactose at 1.6 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, PA-I GALACTOPHILIC LECTIN, ... | | Authors: | Cioci, G, Mitchell, E, Gautier, C, Wimmerova, M, Perez, S, Gilboa-Garber, N, Imberty, A. | | Deposit date: | 2003-07-28 | | Release date: | 2003-12-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of Calcium and Galactose Recognition by the Lectin Pa-Il of Pseudomonas Aeruginosa
FEBS Lett., 555, 2003
|
|
1UOJ
 
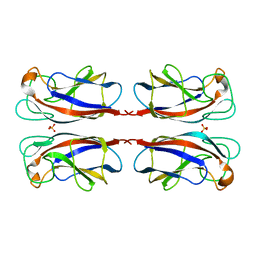 | | CRYSTAL STRUCTURE OF PSEUDOMONAS AERUGINOSA LECTIN 1 IN THE CALCIUM-FREE STATE | | Descriptor: | PA-I GALACTOPHILIC LECTIN, SULFATE ION | | Authors: | Cioci, G, Mitchell, E, Gautier, C, Wimmerova, M, Perez, S, Gilboa-Garber, N, Imberty, A. | | Deposit date: | 2003-09-19 | | Release date: | 2003-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Calcium and Galactose Recognition by the Lectin Pa-Il of Pseudomonas Aeruginosa
FEBS Lett., 555, 2003
|
|
2XRH
 
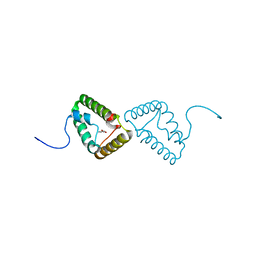 | | Crystal structure of the truncated form of HP0721 | | Descriptor: | NICOTINIC ACID, PROTEIN HP0721 | | Authors: | Cioci, G, Terradot, L, Dian, C, Muller-Dieckmann, C, Leonard, G. | | Deposit date: | 2010-09-15 | | Release date: | 2011-09-28 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Hp0721, a Novel Secreted Protein from Helicobacter Pylori.
Proteins, 79, 2011
|
|
6T1P
 
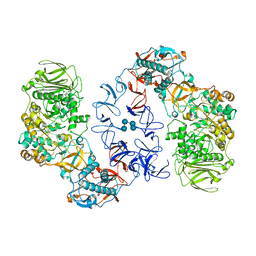 | | ASR Alternansucrase in complex with isomaltononaose | | Descriptor: | Alternansucrase, CALCIUM ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose, ... | | Authors: | Cioci, G, Molina, M, Moulis, C, Remaud-Simeon, M. | | Deposit date: | 2019-10-04 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A specific oligosaccharide-binding site in the alternansucrase catalytic domain mediates alternan elongation.
J.Biol.Chem., 295, 2020
|
|
6T16
 
 | | ASR Alternansucrase in complex with panose | | Descriptor: | Alternansucrase, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Cioci, G, Molina, M, Moulis, C, Remaud-Simeon, M. | | Deposit date: | 2019-10-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A specific oligosaccharide-binding site in the alternansucrase catalytic domain mediates alternan elongation.
J.Biol.Chem., 295, 2020
|
|
6SZI
 
 | | ASR Alternansucrase in complex with isomaltose | | Descriptor: | Alternansucrase, CALCIUM ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Cioci, G, Molina, M, Moulis, C, Remaud-Simeon, M. | | Deposit date: | 2019-10-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A specific oligosaccharide-binding site in the alternansucrase catalytic domain mediates alternan elongation.
J.Biol.Chem., 295, 2020
|
|
6SYQ
 
 | | ASR Alternansucrase in complex with isomaltotriose | | Descriptor: | Alternansucrase, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Cioci, G, Molina, M, Moulis, C, Remaud-Simeon, M. | | Deposit date: | 2019-09-30 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A specific oligosaccharide-binding site in the alternansucrase catalytic domain mediates alternan elongation.
J.Biol.Chem., 295, 2020
|
|
6T18
 
 | | ASR Alternansucrase in complex with oligoalternan | | Descriptor: | Alternansucrase, CALCIUM ION, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose, ... | | Authors: | Cioci, G, Molina, M, Moulis, C, Remaud-Simeon, M. | | Deposit date: | 2019-10-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A specific oligosaccharide-binding site in the alternansucrase catalytic domain mediates alternan elongation.
J.Biol.Chem., 295, 2020
|
|
2BWR
 
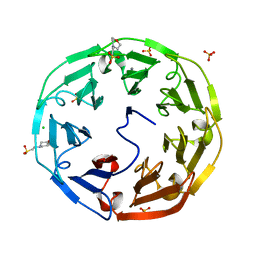 | | Crystal Structure of Psathyrella Velutina Lectin at 1.5A Resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Cioci, G, Mitchell, E.P, Chazalet, V, Gautier, C, Oscarson, S, Debray, H, Perez, S, Imberty, A. | | Deposit date: | 2005-07-18 | | Release date: | 2006-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Beta-Propeller Crystal Structure of Psathyrella Velutina Lectin: An Integrin-Like Fungal Protein Interacting with Monosaccharides and Calcium.
J.Mol.Biol., 357, 2006
|
|
2C4D
 
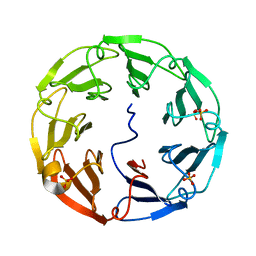 | | 2.6A Crystal Structure of Psathyrella velutina Lectin in Complex with N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PSATHYRELLA VELUTINA LECTIN PVL, ... | | Authors: | Cioci, G, Mitchell, E.P, Chazalet, V, Gautier, C, Oscarson, S, Debray, H, Perez, S, Imberty, A. | | Deposit date: | 2005-10-18 | | Release date: | 2006-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Beta-Propeller Crystal Structure of Psathyrella Velutina Lectin: An Integrin-Like Fungal Protein Interacting with Monosaccharides and Calcium.
J.Mol.Biol., 357, 2006
|
|
2C25
 
 | | 1.8A Crystal Structure of Psathyrella velutina lectin in complex with N-acetylneuraminic acid | | Descriptor: | CALCIUM ION, N-acetyl-alpha-neuraminic acid, PSATHYRELLA VELUTINA LECTIN PVL, ... | | Authors: | Cioci, G, Mitchell, E.P, Chazalet, V, Gautier, C, Oscarson, S, Debray, H, Perez, S, Imberty, A. | | Deposit date: | 2005-09-26 | | Release date: | 2006-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Beta-Propeller Crystal Structure of Psathyrella Velutina Lectin: An Integrin-Like Fungal Protein Interacting with Monosaccharides and Calcium.
J.Mol.Biol., 357, 2006
|
|
2BWM
 
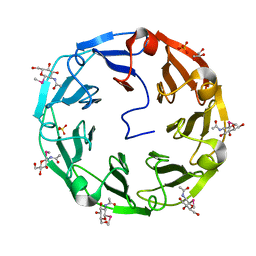 | | 1.8A CRYSTAL STRUCTURE OF of Psathyrella velutina LECTIN IN COMPLEX WITH METHYL 2-ACETAMIDO-1,2-DIDEOXY-1-SELENO-BETA-D-GLUCOPYRANOSIDE | | Descriptor: | CALCIUM ION, GLYCEROL, PSATHYRELLA VELUTINA LECTIN PVL, ... | | Authors: | Cioci, G, Mitchell, E.P, Chazalet, V, Gautier, C, Oscarson, S, Debray, H, Perez, S, Imberty, A. | | Deposit date: | 2005-07-15 | | Release date: | 2006-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Beta-Propeller Crystal Structure of Psathyrella Velutina Lectin: An Integrin-Like Fungal Protein Interacting with Monosaccharides and Calcium.
J.Mol.Biol., 357, 2006
|
|
5LK5
 
 | |
4UDJ
 
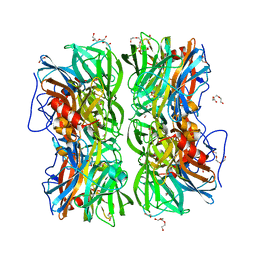 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.60 Angstrom in complex with beta-D-mannopyranose and inorganic phosphate | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UDK
 
 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.76 Angstrom from unknown human gut bacteria (Uhgb_MP) in complex with N-acetyl-D-glucosamine, beta-D-mannopyranose and inorganic phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UDG
 
 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.60 Angstrom in complex with N-acetylglucosamine and inorganic phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5LFC
 
 | | Crystal structure of Leuconostoc citreum NRRL B-1299 N-terminally truncated dextransucrase DSR-M | | Descriptor: | CALCIUM ION, DsrV, GLYCEROL | | Authors: | Claverie, M, Cioci, G, Remaud-simeon, M, Moulis, C, Tranier, S. | | Deposit date: | 2016-07-01 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Investigations on the Determinants Responsible for Low Molar Mass Dextran Formation by DSR-M Dextransucrase
Acs Catalysis, 7, 2017
|
|
4UDI
 
 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.85 Angstrom from unknown human gut bacteria (Uhgb_MP) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6HTV
 
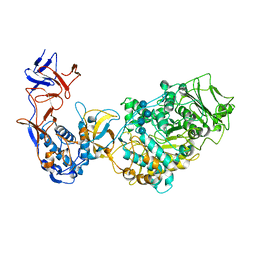 | | Crystal structure of Leuconostoc citreum NRRL B-1299 N-terminally truncated dextransucrase DSR-M in complex with isomaltotetraose | | Descriptor: | Alternansucrase, CALCIUM ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Claverie, M, Cioci, G, Remaud-Simeon, M, Moulis, C, Lippens, G. | | Deposit date: | 2018-10-04 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Futile Encounter Engineering of the DSR-M Dextransucrase Modifies the Resulting Polymer Length.
Biochemistry, 58, 2019
|
|
4UOU
 
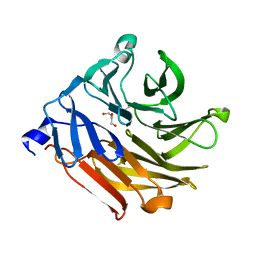 | | Crystal Structure of Fucose binding lectin from Aspergillus Fumigatus (AFL) - apo-form | | Descriptor: | DI(HYDROXYETHYL)ETHER, FUCOSE-SPECIFIC LECTIN FLEA | | Authors: | Houser, J, Komarek, J, Cioci, G, Varrot, A, Imberty, A, Wimmerova, M. | | Deposit date: | 2014-06-10 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights Into Aspergillus Fumigatus Lectin Specificity: Afl Binding Sites are Functionally Non-Equivalent.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5HCB
 
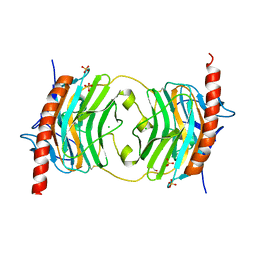 | | Globular Domain of the Entamoeba histolytica calreticulin in complex with glucose | | Descriptor: | CALCIUM ION, CHLORIDE ION, Calreticulin, ... | | Authors: | Moreau, C.P, Cioci, G, Ianello, M, Laffly, E, Chouquet, A, Ferreira, A, Thielens, N.M, Gaboriaud, C. | | Deposit date: | 2016-01-04 | | Release date: | 2016-08-31 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of parasite calreticulins provide insights into their flexibility and dual carbohydrate/peptide-binding properties.
IUCrJ, 3, 2016
|
|
5NGY
 
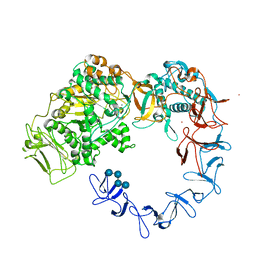 | | Crystal structure of Leuconostoc citreum NRRL B-1299 dextransucrase DSR-M | | Descriptor: | CALCIUM ION, DSR-M glucansucrase inactive mutant E715Q, PRASEODYMIUM ION, ... | | Authors: | Claverie, M, Cioci, G, Remaud-simeon, M, Moulis, C, Tranier, S, Vuillemin, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Investigations on the Determinants Responsible for Low Molar Mass Dextran Formation by DSR-M Dextransucrase
Acs Catalysis, 2017
|
|
6HVG
 
 | |
5O8L
 
 | | Crystal structure of Leuconostoc citreum NRRL B-1299 N-terminally truncated dextransucrase DSR-M in complex with sucrose | | Descriptor: | Alternansucrase, CALCIUM ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Claverie, M, Cioci, G, Remaud-simeon, M, Moulis, C, Tranier, S. | | Deposit date: | 2017-06-13 | | Release date: | 2017-11-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Investigations on the Determinants Responsible for Low Molar Mass Dextran Formation by DSR-M Dextransucrase
Acs Catalysis, 7, 2017
|
|
2XR4
 
 | | C-terminal domain of BC2L-C Lectin from Burkholderia cenocepacia | | Descriptor: | LECTIN, SULFATE ION | | Authors: | Sulak, O, Cioci, G, Lameignere, E, Delia, M, Wimmerova, M, Imberty, A. | | Deposit date: | 2010-09-09 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Burkholderia Cenocepacia Bc2L-C is a Super Lectin with Dual Specificity and Proinflammatory Activity.
Plos Pathog., 7, 2011
|
|
